Systems featuring programmable RNA molecule export capabilities hold the potential for non-destructive cellular dynamics monitoring and efficient RNA delivery to engineered cells. In a recent breakthrough, Michael B. Elowitz and Felix Horns from Caltech presented their findings in the journal Cell, unveiling virus-inspired genetically encoded cellular RNA exporters. These exporters efficiently package and secrete cargo RNA from mammalian cells while safeguarding them within nanoparticles. Through RNA barcode exportation and sequencing, these systems facilitate the non-destructive monitoring of cell population dynamics at a clonal level. Additionally, by incorporating fusion proteins into nanoparticles, they demonstrate the delivery, expression, and functional activity of exported mRNA in recipient cells. These innovative systems are referred to as COURIER (controlled output and uptake of RNA for interrogation, expression, and regulation), paving the way for measuring cellular dynamics and advancing hybrid cell and gene therapies grounded in intercellular RNA delivery.

RNA, as the central information carrier in cells, serves as a potent “interface” for observing and influencing cellular behavior. Analyzing RNA through sequencing provides insight into a cell’s state, while RNA expression exerts control over cellular status. Unfortunately, RNA is often confined within the cell of its origin, limiting its utility for molecular analysis and intercellular communication. In contrast, the ability to programmably export RNA molecules from cells opens up avenues for analyzing and manipulating living cells.
RNA export/secretion offers a non-destructive means of monitoring cellular dynamics. Techniques like single-cell RNA sequencing and hybrid genomics have transformed the biomedical field, enabling us to decipher the molecular characteristics and states of individual cells. However, accessing RNA from living cells often requires cell lysis or fixation, preventing the tracking of dynamic behavior. On the other hand, sequencing cell-free RNA, which cells naturally secrete through extracellular vesicles (EVs) or upon death, can nondestructively reveal biomarkers of health and disease. However, the limited rate of natural RNA secretion restricts the sensitivity and information content of cell-free RNA analysis. An alternative approach involves engineering cells to efficiently export RNA molecules that encode information about cell populations and states, then collecting and sequencing these secreted RNAs. This method provides unparalleled insights into cell dynamics compared to natural cell-free RNA analysis, offering destructive measurements with increased sensitivity and information content.
RNA export/secretion also opens up possibilities for manipulating cellular behavior. RNA’s capacity to encode proteins and regulate gene expression enables programmable control over cell behavior. However, the therapeutic utilization of this capability is hindered by the challenge of delivering RNA to specific cell populations within tissues. Engineering cells to export RNA presents the opportunity to create therapeutic “delivery cells” that home to tissues, recognize target cells, and perform various RNA-based functions within recipient cells. These functions include altering gene expression, reprogramming cell fate, or selectively eliminating cells in disease states. This approach circumvents the challenges associated with other delivery vehicles, as cells can penetrate tissues and employ cell-based sensing and logic to conditionally regulate localized RNA delivery. A pivotal element of this vision is a system capable of secreting RNA through a vector, allowing non-engineered recipient cells to take up and express these RNAs.
Virus-like particles (VLPs) and exosomes (EVs) emerge as promising platforms for RNA secretion and delivery. Exploiting viral structural proteins and their inherent interactions with RNA packaging signals (PSs), researchers have harnessed VLPs to package and transfer RNA between cells. However, these methods often rely on retroviral coat proteins, such as those of HIV or MMLV, which exhibit limited binding specificity for viral RNA and readily bind other RNAs, posing challenges for selective payload RNA loading. Modifications such as fusing RNA-binding proteins into the coat or incorporating proteins into EVs, along with tagging payload RNA with homologous interacting sequences, have been implemented to enhance selective payload RNA loading. These methods have been used for RNA delivery, including in mice, but require further refinement due to issues such as inefficient payload loading and secretion, limited payload capacity and poor payload expression post-delivery, coat modifications that affect VLP assembly, etc.
An optimal RNA secretion system must address several critical requirements:
- Efficient secretion of RNA from mammalian cells for precise measurement and potent delivery.
- Selective export of target RNA, including engineered barcodes or cargo.
- Protection of exported RNA from extracellular RNase degradation.
- Enablement of cargo RNA delivery and expression in recipient cells. Minimal disruption to the expressing cells when system components are expressed.
An RNA secretion export system with these features serve as create a versatile RNA-based reporter and delivery platform. This study reports the development of RNA secretion export systems embodying these characteristics and their application for non-destructively monitoring cell population dynamics and facilitating intercellular mRNA delivery. Our approach involves designing a set of RNA secretion exporters, which integrate three modular protein components:
(1) RNA binding proteins for specific RNA molecule capture.
(2) Self-assembled shells or vesicles to package and secrete these RNAs.
(3) Fusionin for the targeted delivery of secreted RNA to target cells.
Our journey begins with VLPs, progressing through several generations of RNA exporter engineering, culminating in the development ofprotein nanocage-based extracellular vesicles. These vesicles efficiently package and secrete RNA from cells while precisely targeting RNA step-by-step. Furthermore, we combine RNA export with genetic barcoding and sequencing to non-destructively monitor cell population dynamics. Finally, we demonstrate the delivery and functional activity of cargo RNA, including mRNA encoding Cre recombinase and fluorescent proteins, into target cells by incorporating fusion protein into secreted nanoparticles. The authors term these systems COURIER for controlling RNA export and uptake for interrogation, expression, and regulation. These results establish COURIER as a flexible and scalable paradigm for non-destructive cellular dynamics measurement and intercellular RNA transfer.
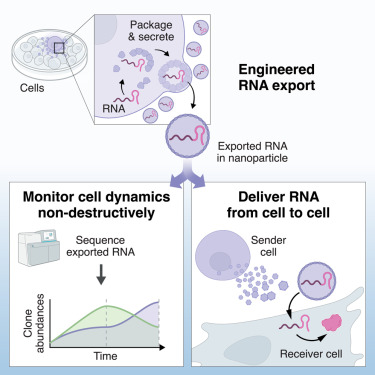 Development of RNA exporters, based on shells and nanocages, for packaging and secretion of RNA, non-destructive monitoring of cells, and cell-to-cell delivery of mRNA.
Development of RNA exporters, based on shells and nanocages, for packaging and secretion of RNA, non-destructive monitoring of cells, and cell-to-cell delivery of mRNA.
References:
Horns F, Martinez JA, Fan C, Haque M, Linton JM, Tobin V, Santat L, Maggiolo AO, Bjorkman PJ, Lois C, Elowitz MB. Engineering RNA export for measurement and manipulation of living cells. Cell. 2023 Aug 17;186(17):3642-3658.e32. doi: 10.1016/j.cell.2023.06.013. Epub 2023 Jul 11. PMID: 37437570.
Related Services:
RNA Isolation and Profiling Services for Extracellular Vesicle
