Introduction
Single domains represent the smallest known fragment still capable of binding antigen that can be isolated from a full-sized immunoglobulin. In work on the cloning of antibody genes and construction of human antibody libraries, Greg Winter and his colleagues found that single domains comprised of either VH or VL derived from human antibodies could also be stabilized as stand-alone, antibody fragments. Shortly thereafter, heavy-chain-only antibodies (HCAbs) were discovered to occur naturally in camelids where the binding domains constitute paired VHs with no light chains. HCAbs are thought to represent up to 50–80% of the antibody repertoire in camels and up to 10–25% of the antibody repertoire in other members of the Camelidae family such as llamas. Similar antibodies also lacking a CH1 domain and a light chain termed Ig-NAR have also been identified in nurse sharks, although these will not be discussed further in this overview (Figure 1). More recently, a new type of “domain antibody” was constructed using the CH2 domain of an IgG as the base scaffold into which CDR loops were grafted. These antibodies with unpaired antigen binding domains, and the single domain versions derived from them, have unique properties and have already shown promise as potential therapeutic antibodies. For the purposes of this review, these molecules (whether derived from VH or VL variable regions or isolated from camelids) will be described as single-domain antibodies (sdAbs).
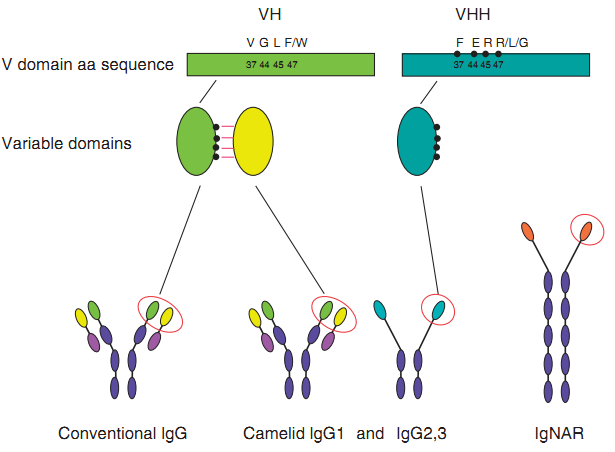
Figure 1 Single-domain antibodies(sdabs). Sdabs can be derived from either conventional mAbs or from HCAbs or IgNARs. Camelid VHs andVHHs differ in four hallmark amino acid residues found in the VH/VL interface. (de Marco, A. 2011)
Since these initial researches, there has been an explosion of interest aimed at understanding the unique properties of sdAbs. The primary advantages of sdAbs as compared with scFvs are generally better folding and stability characteristics, the absence of the linker, and size. Table 1 compares some of the high-level similarities and differences of Fab fragments, scFvs, and single domain antibodies. With a molecular weight of around 15 kDa, sdAbs are amenable to applications that require enhanced tissue penetration or rapid clearance, such as radioisotope-based imaging. Because of their small size, sdAbs are below the renal clearance cutoff, with a resulting half-life that is significantly shorter than full-sized mAbs. This can be extended using modifications such as PEGs (polyethylene glycols), by Fc-fusion, or by binding to long-lived serum components, for example, albumin. Fc-fusion has the additional benefit of allowing the exploitation of mAb-like properties such as sdAb-directed antibody-dependent cell-mediated cytotoxicity (ADCC) and FcRn recycling. In addition, sdAbs allow higher molar doses for the same (mg kg−1) amount when compared to monoclonal antibodies.
Table 1 Comparison of general properties of different forms of antibody fragments
| Property | FAb fragments | ScFvs | sdAbs ** |
| Size | ~50 kDa (FAbs) | ~25 kDa | ~12 kDa |
| Proven commodity | +++ (severalmarketed) | +/– (late phaseClinical trials) | +/– (early phase clinical trials) |
| High affinity | + | +/– | +/– (depends ontechnology used) |
| Manufacturing | CHO, E. coli, yeasts | E. coli, yeasts | E. coli |
| Stability (e.g. pH, tempera xidation, shear stress) | + | +/– | ++ |
| Ease of purification | +/– | +/– | +/– |
| Potential for ambient temperaturestorage formulation | +/– | – | + |
| Alternative routes ofadministration (e.g. intranasal,inhalation) | – | +/– | + |
| Potential for blocking enzyme active sites | +/– | +/– | + (particularly nanobodies and IgNARs) |
| Tissue penetration | ++ | +++ | ++++ |
| Cavity binding | – | – | ++ |
| Manufacturable bi-,tri-,and multi-specificity | – | +/– | ++ |
| Effector functionality (e.g. ADCC,ADPC, CDC) | – | – | – |
| Serum half-life (without half-lifeextension technology) | <1 h | < 1 h | <1 h*** |
| Ability to extend half-life | +++ (up to ~10–12d with PEGylation)* | +++ (up to ~10–12 d with PEGylation)* | +++ (up to ~10–12 d with PEGylation)*,*** |
| Potential for intracellular targeting | – | +/– | + (especially if disulfide bondsare removed) |
| Ease of manipulation | + | +/– (some aggregation issues) | + |
| Ability to dimerize or concatenate | – | + | ++ |
**General principles covering both human domain antibodies, camelid sdAbs, and IgN antibodies.
***Ability to make albumin binding domains that can be fused to domain antibodies, yielding better half-life with only modest increase in size.
Generation of sdAbs
Single domain antibodies are generally either isolated from Camelidae or based on human frameworks. A diverse sdAb (VHH) library can be prepared in two methods: by amplification of VHH genes from isolated lymphocytes of naïve or immunized members of Camelidae, or by introducing diversity into a VHH scaffold synthetically. Libraries of human sdAbs (VK or VH) are mainly based on synthetic libraries where diversity is introduced into one or more scaffolds.The recent generation of rodents expressing human HCAbs, devoid of rodent immunoglobulin chains, now provides an alternative route for isolating human single VH (but not VL) domains.
Isolating sdAbs from immune camelid libraries is a popular way over the last decade. The most commonly used method involves the immunization of a member of the Camelidae family with the antigen of interest, recovery of lymphocytes from the immunized animal, preparation of the cDNA, generation of a phage display library using standard cloning protocols, and finally, three to four rounds of phage screening to enrich antigen-specific binders. Because of the naturally occurring affinity maturation process of antibodies in vivo, driven by somatic hypermutation, the repertoires of sdAbs obtained from immunized animals frequently contain a large proportion of high-affinity binders to the antigen used for immunization. Antigen-specific sdAbs can be also selected from naïve libraries isolated from nonimmunized animals which can be used for selections against multiple antigens. This approach is often at the cost of affinity, yielding clones with lower affinities to the cognate antigen because the library consists of naïve antibodies that have not undergone affinity maturation in vivo. The majority of the diversity present in camelid-derived sdAb libraries is generated in vivo; by contrast, the diversity present in synthetic libraries is achieved through the use of diversifying oligonucleotides in conjunction with combinatorial approaches. Synthetic libraries can be of use when there is a need for a sdAb against a target that is poorly immunogenic, for example, amyloid β fibrils. Diversity can be introduced into the human antibody scaffold through the use of both diversifying oligonucleotides and recombination of CDRs or by grafting CDRs from pre-existing naïve or immune human Fab antibody library repertoires. This allows the identification of specific binders with low nanomolar affinities to components of insulin-like growth factor (IGF) system that had not been screened from an earlier library based on this framework. Several approaches have been taken to generate human sdAbs that demonstrate both high affinity and good biophysical properties, since the stability of human sdAbs is variable and depends on both the framework used and the CDR sequences. Some organizations have developed transgenic mice and rats capable of expressing fully functional human HCAbs. Advances in methods of stably introducing artificial chromosomes into rodents, combined with silencing the endogenous IgG loci, have proved capable of allowing the generation of mice and rats that can produce these functional human sdAbs.
Whatever the method used to generate the library or whether the camelid/human sdAb library, identification of a target-specific antibody suitable for therapeutic use needs in vitro screening and characterization. The most traditional method to enrich for antigen-specific binders from either camelid or human sdAb libraries is phage display, although a number of other display methods have been successfully investigated, including ribosome, yeast, and bacterial display. Phage display involves fusing libraries of sdAb genes to a phage coat protein and displaying the sdAbs on phage particle surface. The majority of sdAbs have been isolated in this way, owing to the relative simplicity and the ability to rapidly enrich binders of this technique. One potential shortcoming of phage display is the variable levels of sdAb display on the phage particle surface, which can lead to faster enrichment of clones with lower affinity but higher display levels. Alternative selection systems to phage display include both bacteria and yeast display, which are facilitated by fluorescent-activated cell sorting (FACS). SdAb affinity can be directly measured by using FACS, while it is still displayed on the cell surface can allow discrimination between clones differing by only a small degree in affinity. Another advantage of using yeast display is that the secretory pathways in yeast are similar to those of higher eukaryotes. However, the major limitation of sdAb display on the yeast has been its low transformation efficiency. All of the display techniques described above involve a cell transformation step, the efficiency of which limits the size of the library. Alternative display techniques hat do not require a cell transformation step, such as ribosome display, is viable alternatives to cell-based systems.
Outlook
Recently, a range of high-affinity sdAbs of both camelid and human origin have been isolated by in vivo and in vitro approaches against a broad scope of targets. These sdAbs have been found to applicate in both the therapeutic and diagnostic fields. Monomeric sdAbs have value as diagnostic tools and also therapeutically both in imaging, where their short life aids the production of high contrast images. sdAbs can also form the basis of a number of bi-specific formats either utilizing multiple sdAbs in tandem or through fusion to a more conventional mAb framework. sdAbs targeting tissue or cell-specific markers can be used to target payloads such as peptides, small molecules, and oligonucleotides.
The modularity, good expression, and biophysical properties of sdAbs makes them an attractive option in exploring this broad range of formats and will aid their development from early stage clinical assets into medicines of value in the future.
