Proteolysis targeting chimera (Protein Degrader) technology represents a breakthrough development in the field of drug discovery, utilizing the ubiquitin proteasome system to specifically degrade disease-related proteins. Protein Degraders are characterized by their unique heterobifunctional structure, including two functional domains connected via linkers.
Linker plays a key role in determining the biodegradable efficacy of Protein Degraders. An advanced and well-designed Protein Degrader functional linker is under development. Despite this, the correlation between linker characteristics and Protein Degrader efficacy remains understudied.
We mainly analyze and discuss the structure types and characteristics of the Protein Degrader linker, its reasonable design and optimization strategy, and the influence of linker characteristics on the biodegradation efficiency of Protein Degrader.
Structure types and characteristics of Protein Degrader linker
Protein Degrader linkers can be broadly divided into flexible linkers and relatively rigid linkers.
Flexible linkers are the most widely used type, mainly alkyl linker and polyethylene glycol (PEG) linkers. The flexible linkers that are ubiquitous in Protein Degrader designs typically use long, flexible structures. However, this design can increase susceptibility to oxidative metabolism in the body.
In contrast, due to the limitations of synthesis technology in the field of Protein Degrader, the relatively rigid linker is used less frequently, including a total of 8 types. Among them, naphthenes, especially linkers containing piperazine and piperidine components, are often used because they increase the solubility and stability of ternary complexes. Triazole-based linker is another commonly used rigid linker that uses copper-catalyzed 1,3-dipole cycloaddition reactions with high compatibility and consistent reaction rates with different functional groups due to the advent of click chemistry. Another type of linker, the photocontrolled Protein Degrader linker, such as the photoswitch Protein Degrader linker, uses azo fragments instead of alkyl or polyether fragments, which, when exposed to specific wavelengths of light, result in reversible photoisomerization of the resulting Protein Degraders. This makes it possible to precisely and mutually adjust the biodegradation rate of Protein Degraders (Figure 1).
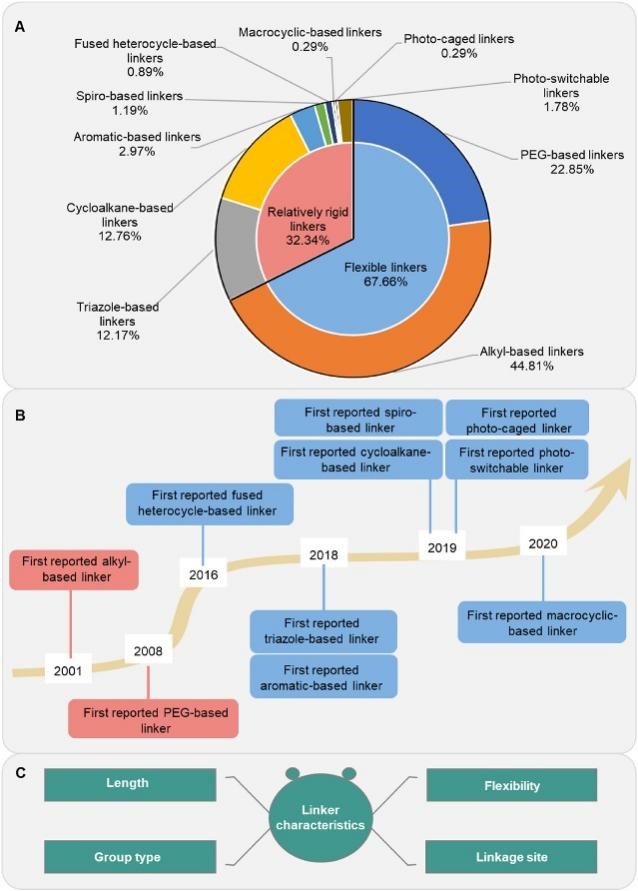
Fig. 1 Structural types and characteristics of Protein Degrader linkers.1
Rational design and optimization strategy of Protein Degrader linker
In general, the design and optimization of Protein Degrader linkers rely on empirical or computer-aided methods. The general journey of designing and optimizing a Protein Degrader linker based on experience is as follows:
First, the researchers designed Protein Degraders based on experience and then focused on optimizing linkers in four areas:
- Adjust the linker length to achieve the optimal configuration of a particular Protein Degrader.
- Modify the type of linker to balance the hydrophilicity and hydrophobicity of Protein Degraders.
- Modify the flexibility of the linker and junction sites to increase the stability of the ternary complex.
- Design and synthesize multiple Protein Degraders with different linkers.
Whereas, the general process based on computer-aided strategies is:
- Use crystallography or molecular docking techniques to determine the binding mode of POI to its ligand and E3 to its ligand, respectively.
- Global protein-protein docking simulations were carried out by computational software such as MOE, Rosetta, and PatchDock to obtain model structure sets of poi-ligand and e3-ligand.
- After evaluation using molecular dynamics (MD), analyze protein-protein interactions (PPIs) using a rational model structure and identify proteins that interact proximal to their binding pockets.
- Design different linkers to generate a series of POI-Protein Degrader-E3 conformations. The structure-activity relationship (SAR) and binding mode of the ternary complex were further analyzed, and the optimal linker was finally obtained.
This strategy is widely applicable to the design and optimization of Linker, however, this approach has certain limitations. This approach often requires the synthesis of multiple protacs with various linkers to gain a comprehensive understanding of SAR, which can be time-consuming, laborious, and costly. Another limitation is the accuracy of theoretical predictions, which affects the design of the linker.
In recent years, the application of artificial intelligence in the design of Protein Degrader joints has significantly improved the accuracy and efficiency of linker optimization. By leveraging AI strategies, researchers have gained insight into the structure, physics, and chemistry of the PORTAC linker. These insights have been translated into real-world applications, leading to more precise Protein Degrader linker designs (Figure 2).
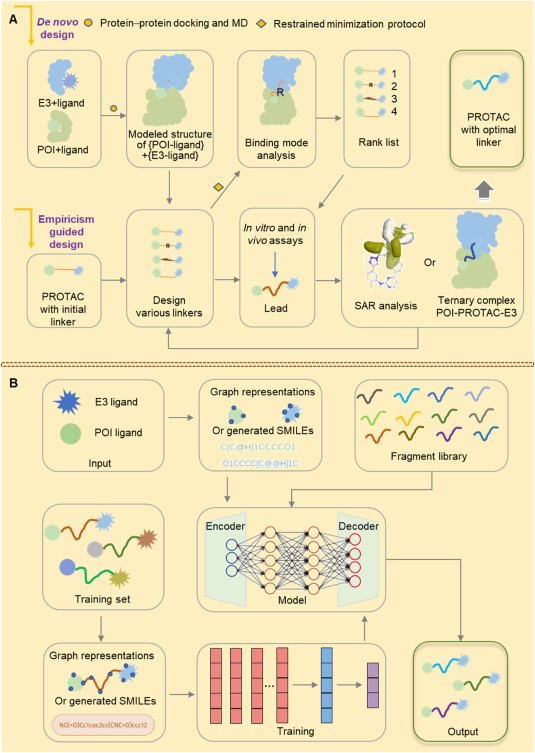
Fig. 2 Appropriate design and optimization strategies for Protein Degrader linkers.1
Effect on the biodegradation efficiency of Protein Degraders
Protein Degrader is a heterobifunctional molecule linked by a target protein ligand and an E3 ligase ligand via different linkers, and this structure aids in the establishment of a stable ternary complex, which allows the organism’s 26s proteasome to recognize and degrade the target protein ubiquitination.
It was found that the properties of linkers play a key role in the stable formation of ternary complexes and the physicochemical and pharmacokinetic properties of Protein Degraders.
Firstly, the length of the linker significantly affects the formation of the POI-Protein Degrader-E3 ternary complex, and the optimal linker length depends on the interaction mode, distance, and spatial structure of the ternary complex. On the other hand, shorter linkers may introduce steric hindrance, disrupt the formation of ternary complexes, and reduce the biodegradation efficiency of Protein Degraders.
Secondly, the chemical composition of the linker directly affects the physicochemical properties of Protein Degraders, which in turn affect the cellular permeability of Protein Degraders, thereby significantly affecting the biodegradation efficiency of Protein Degraders.
Therefore, achieving optimal linker length is critical to generating maximum interaction between POI and E3 ligase, resulting in efficient ubiquitination and biodegradation of POI. On the other hand, the flexibility of the linker is a key factor in determining the biodegradation effect of Protein Degraders, and a linker with significant conformational flexibility can enhance the interaction between Protein Degraders, POIs, and E3 proteins, thereby preventing their stable binding at the fixed interface.
On the contrary, the introduction of rigid groups in the flexible linker can improve Protein Degrader rigidity and replicate the original geometry of POI and E3 ligands in Protein Degraders, resulting in new interactions and improved stability of ternary complexes.
In addition, the linker’s attachment site to the POI and E3 affects the interaction between the POI and E3. Optimization of the Protein Degrader linker typically involves identifying the most favorable site for structural derivatization, thus ensuring that maximum binding affinity is preserved. This selection process typically involves analyzing the solvent-exposed region of the POI-ligand or E3-ligand interaction interface, and by introducing an optimal linker in the solvent-exposed region, the protein-protein interaction can be maximized while preserving the original ligand interaction with the POI or E3.
Given these intricate relationships, it is clear that the linker is a key factor in ensuring greater specificity and targeting efficiency of Protein Degraders.
While optimizing the length, group type, flexibility, and junction site of the linker can improve the efficacy of Protein Degraders, several challenges must be overcome.
- The complexity and diversity of Protein Degrader structures hinder the establishment of clear structure-activity relationships. To address this issue, exploring the optimal linker characteristics of specific biodegradable systems can facilitate the rapid identification of more effective Protein Degraders.
- The physicochemical properties of Protein Degraders often deviate from the classical drug-like 5 principle, and it is necessary to study the drug similarity rules suitable for Protein Degraders to minimize the possibility of designing Protein Degraders with poor pharmacokinetic characteristics.
- Due to their large molecular weight and complex structure, purification and low yield pose challenges to the synthesis and optimization of Protein Degraders. Therefore, the development of advanced synthesis techniques for Protein Degraders is essential to obtaining a wider range of structure types.
- The large size of the ternary complex composed of POI-Protein Degrader-E3 makes the determination of crystal structure difficult. To solve this problem, it is necessary to develop advanced crystallography techniques or more accurate computational simulation methods to gain insights for further optimization.
Solving these problems will greatly promote the progress of Protein Degrader research.
References
- Dong, Yawen, et al. “Characteristic roadmap of linker governs the rational design of Protein Degraders.” Acta Pharmaceutica Sinica B(2024).
