Affinity maturation is the process to improve antibody affinity for an antigen. In vivo, natural affinity maturation by the immune system takes place by somatic hypermutation and clonal selection. In vitro, in the laboratory affinity maturation, can be obtained by mutation and selection.
Creative Biolabs has gained extensive experience in antibody affinity maturation. We usually take scFv as the antibody format in affinity maturation. Also, a monovalent display phagemid system is used to reduce the avidity effects during antigen-binding screening. We also provide affinity maturation services for single domain antibodies. Two methods, untargeted mutagenesis and oligonucleotide-directed mutagenesis, are employed to construct random or defined sub-libraries to introduce a large number of mutants of the original antibody. Antibody binders of higher affinity are then selected by increasing the screening stringency. By constructing a series of sub-libraries of a scFv/Fab antibody, our proprietary protocol allows increase of the affinity of the scFv antibodies from 10-9 to 10-10. We have successfully obtained a scFv antibody that has an extremely high affinity of 10-12, whose binding to the antigen is essentially irreversible.

We use an error-prone PCR integrated DNA-shuffling approach to mutate mainly CDR regions during sub-library construction. If the potential of introducing immunogenic mutations to framework positions is not a concern, we usually use this approach to create mutations at completely random positions across the entire VH and VL fragments. In these cases, the genetic diversity of the sub-library is further increased via passage through our proprietary bacterial mutator strain, CD-affi™.
If the structure of the antibody/antigen complex is available or modeling the structure of the antibody/antigen is possible, certain positions can be randomized at a defined diversity (such as full randomization with all 20 amino acids or biased randomization with selected amino acids at fixed percentages) to improve the affinity. We are able to create any sub-libraries to incorporate the defined mutations using trimer codon technology. Most of the time, we need study the AA sequences of the antibody to find out the conserved sequences (in comparison with the germ-line and antibody subfamily sequences). We may then introduce mutations to the positions in the frame work regions that are not conserved. Supposedly, these regions will be antigen-specific and change in these regions may not increase immunogenicity.
Subsequent library screening will fish out the antibody mutants that have high affinity. Two library screening strategies are available. In the first "surface-panning" strategy, decreasing concentrations of antigen is surface immobilized. In the second "solution-sorting" strategy, in which a labeled antigen in solution is used, we have two approaches, selection based on the equilibrium constant (Kd) and selection based on binding kinetics. In the first approach, sub-library phage is incubated with biotinylated antigen at controlled concentrations and bound phages are captured by immobilized NeutrAvidin. Selection based on binding kinetics is also termed off-rate (Koff) selection, in which phage population is allowed to saturate the labeled antigen before a large molar excess of unlabeled antigen is added to the mix for controlled periods of time. This allows the selection of mutant antibodies that have slower off-rates. Since a reduction in Koff usually results in a higher affinity, this selection approach singles out antibody variants with improved Kd.
We offer Biacore Analysis services for binding kinetic analyses of antibodies. We typically capture the antibody on the chip and run antigen on top of the captured antibody. The antigen will be ran at 6 different concentrations for each antibody and chi-square analysis will be performed on the binding constants we obtain from each antigen concentration. The documentation package will include a real time on-rate (Ka), off rate (Kd), an affinity constant (KD), chi square value and a graph of real-time binding kinetics. We would like to obtain ~50 uL of 1 mg/mL antigen and antibody solutions. We will need ~100 ug of antigen and ~50ug for each antibody. We would need MW information for the antigen as well. It may require special considerations for antigens with repeated or multiple epitopes for affinity determination.>> Learn more about Antibody Affinity Measurement Services
Alanine scanning mutagenesis is our favorite method in affinity maturation of peptide binders. In this method, each single AA of a selected binding peptide will be replaced with an alanine, and then the binding of the modified peptides to the target protein will be assayed using Biacore technology. The non-essential AAs will be specifically identified. After that, we will create a directed/constrained peptide sub-library that contains random sequences on the non-essential AA positions. Here, again, we frequently randomize the non-essential residues using "NNK" or "trimer codon" strategy. Mutants with increased binding affinity are identified by enhancing the screening stringency, followed by phage ELISA.
Other optional antibody engineering services:
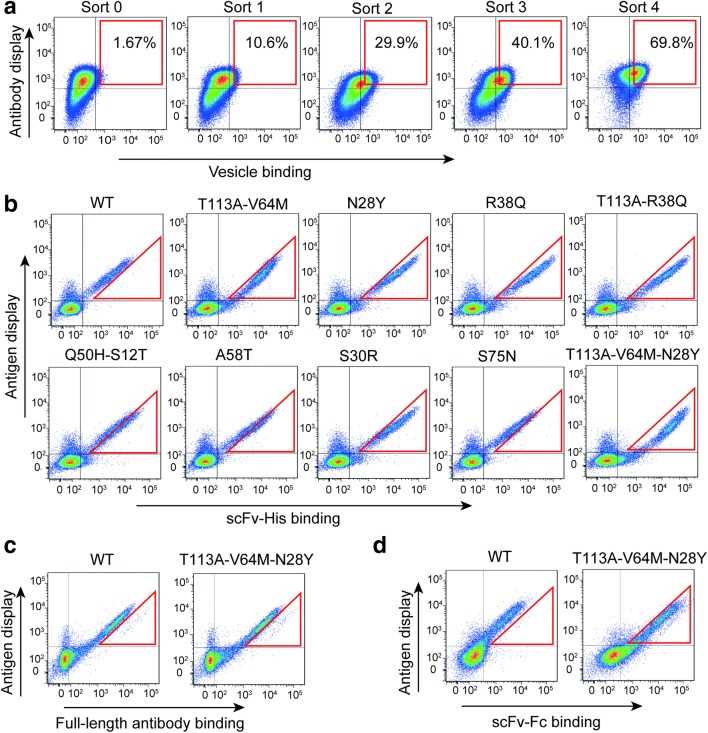 Fig. 2 Affinity maturation of single-chain antibodies against ETaR. (Jie Wang, 2019)
Fig. 2 Affinity maturation of single-chain antibodies against ETaR. (Jie Wang, 2019)
G protein-coupled receptors (GPCRs) are targets for many drug molecules, but developing anti-GPCR drugs faces huge challenges, that is, they are insoluble in aqueous solutions and are difficult to affinity mature. Here, the researchers used CHO cell display libraries of single-chain variable fragments (scFvs) and full-length antibodies to mature directly against vesicle probes prepared from CHO cells displaying the endothelin A receptor (ETaR) GPCR. They used vesicles as probes and combined them with the CHO cell display platform to conduct four rounds of antibody affinity maturation, which increased the affinity of scFv by 13.58 times and the affinity of full-length antibodies by 5.05 times. The researchers said they expect this method to be used not only for affinity maturation of antibodies against GPCRs, but also for maturation of antibodies against other types of proteins.
Antibody affinity maturation is a natural process that occurs in the immune system, enhancing the binding affinity of antibodies for their specific antigens. This process typically takes place in the germinal centers of lymph nodes after initial exposure to an antigen. Through a combination of somatic hypermutation (SHM) of antibody genes and selective clonal expansion, B cells that produce antibodies with higher affinity for the antigen are preferentially selected.
In a laboratory setting, antibody affinity maturation can be mimicked using various techniques such as phage display, yeast display, or mammalian display. These methods involve generating a library of antibody variants with slightly altered sequences. The antibodies are then tested for their ability to bind the target antigen, and those with higher affinity are selected. This selection process is often repeated multiple times to progressively enhance the antibody's affinity.
High-affinity antibodies are crucial for effective therapeutic outcomes as they bind more strongly and specifically to their target, which can improve the efficacy of the antibody in neutralizing pathogens or targeting diseased cells. Enhanced affinity can also allow for lower doses of antibodies to be used, potentially reducing the risk of adverse effects and the cost of therapy.
Higher affinity antibodies often exhibit longer retention times at their target sites, which can prolong their therapeutic effects. This extended duration of action means that treatments might be administered less frequently, enhancing patient compliance and overall treatment efficacy.
There is a potential risk that antibody affinity maturation could lead to autoimmunity. During the maturation process, antibodies may acquire higher affinity for self-antigens, leading to an autoimmune response. This underscores the importance of careful screening and validation processes in the development of therapeutic antibodies to ensure that they do not react with host tissues.
Computational methods can predict how changes in antibody sequences might affect their structure and interaction with antigens. By using computational modeling and simulations, researchers can identify promising candidates for mutagenesis, potentially reducing the number of experimental iterations needed to achieve high-affinity antibodies.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
