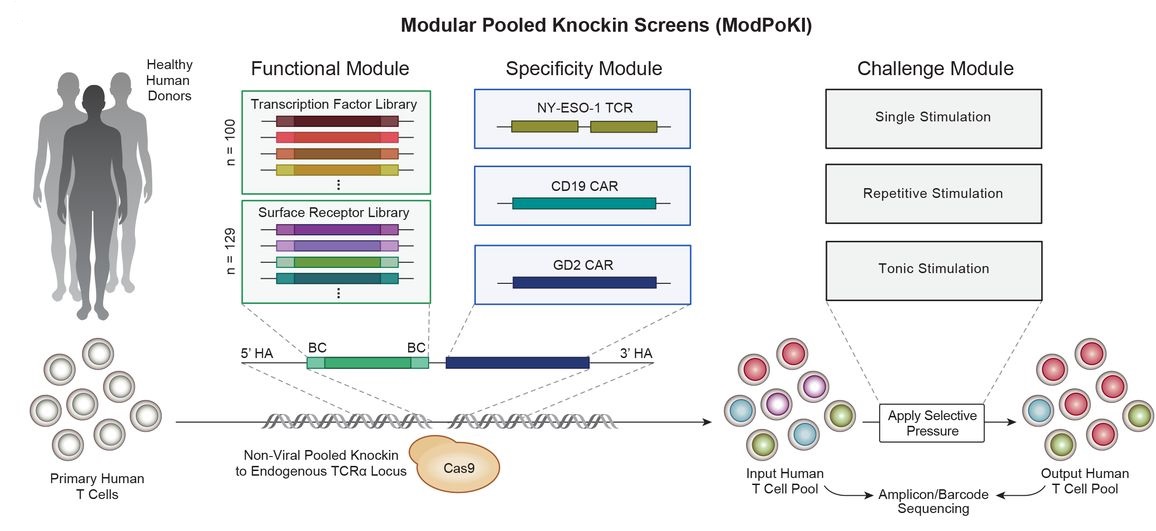
In recent years, scientists have harnessed the power of gene editing technology to reprogram immune cells into therapies capable of targeting cancer. However, these immunotherapies are not universally effective across all patients or types of cancer. Furthermore, identifying the precise combination of genetic changes that can enhance these reprogrammed immune cells is a daunting and time-consuming task.
Now, in a groundbreaking study, researchers from the Gladstone Institutes and the University of California, San Francisco, have developed a technique that allows them to rapidly “assemble” thousands of different gene editing combinations for testing in immune cells. They have utilized a screening technology known as “Modular Pooled Knockin Screening” (ModPoKI) to identify a novel gene combination that, when introduced into immune cells, prolongs their lifespan and enhances their anti-cancer efficacy. The research findings were published in the September 14, 2023, issue of the journal Cell, titled “Modular pooled discovery of synthetic knockin sequences to program durable cell therapies.”
Dr. Alexander Marson, co-corresponding author of the study and Director of the Gladstone-UCSF Institute of Genomic Immunology, stated, “This is a significant step forward for us. We now have the capability to pose questions about how to assemble fragments of genetic programs into cells and test how they benefit patients. I believe this will accelerate the development of improved cell therapies.”
Dr. Ansuman Satpathy, co-author of the study and Assistant Professor in the Department of Pathology at Stanford University School of Medicine, added, “This new research demonstrates the power of using high-throughput genomics to discover and design novel molecular programs in cell therapy, further understanding how these programs impact the T-cell states required to kill cancer.”
Mixing and Matching Components
T cells of the immune system play a crucial role in the anti-cancer process. When T cells recognize cancer cells as foreign entities through receptors on their surface, they target and destroy the cancer cells. Scientists have found ways to modify these receptors on T cells, often by adding DNA sequences encoding chimeric antigen receptors (CARs), to enable T cells to more easily identify and eliminate cancer cells.
In CAR-T cell therapy, T cells are extracted from cancer patients’ blood, genetically reprogrammed in the lab to introduce these new CAR receptors, and then infused back into the patient’s bloodstream. However, many CAR-T cell therapies still have limitations: they often prove ineffective against solid tumors, degrade over time, and some CARs may not elicit a strong enough immune response to kill cancer cells.
Dr. Franziska Blaeschke, the first author of the paper and a postdoctoral researcher in the Marson lab, explained, “CAR-T cells have achieved incredible success in treating blood cancers such as leukemia and lymphoma, but we are still searching for ways to optimize them and apply them to other cancers. Until now, we’ve lacked a systematic approach to discovering which genetic changes in T cells are most effective for improving CAR-T cells.”
To fill this gap, the authors developed ModPoKI. This technique assembles multiple genes into long DNA segments for use in the CRISPR gene editing platform. Using this tool, they created approximately 10,000 potential combinations by mixing hundreds of genes with the DNA encoding specific CARs. Subsequently, they used CRISPR to insert these spliced DNA sequences into a defined location in T cell genomes.
Each T cell received a different DNA sequence, and these cells were then pitted against each other to see which T cell performed best in various predictive tests of anti-tumor activity. Each gene combination generated by ModPoKI had an easily readable DNA barcode, allowing researchers to track which gene combinations improved T cell function.
This Lego-like capability enabled them to quickly discover gene combinations that could enhance CAR-T cells without the need for manual selection and design of each gene combination before introducing them into cells.
Dr. Theodore Roth, co-corresponding author and former member of the Marson lab, said, “We don’t have to individually guess what can improve cell function and then study them one by one; we can put these fragments together and rapidly test many cells. This is a highly valuable aspect of molecular biology.”
Towards Better Therapies
In this new study, the genes added to the cells by the Marson team were surface receptors (both natural and engineered receptors designed to send enhanced signals to CAR-T cells) and transcription factors (genes that activate or deactivate other genes). When analyzing the results of testing hundreds of surface receptors and transcription factors, the authors found that different CARs could be optimized through different factors.
Dr. Blaeschke explained, “It turns out that transcription factors are not one-size-fits-all. Therefore, when scientists are developing new CAR-T cells, it’s essential to check what other factors can optimize CAR-T cells. Our research creates a roadmap that scientists can use to combine different transcription factors with different T cell receptors (TCRs) or CARs.”
The authors also identified two combinations of transcription factors that seemed to consistently enhance CAR-T cells. These two transcription factors, BATF and TFAP4, improved the adaptability of CAR-T cells previously developed for treating pediatric brain tumors.
Dr. Marson noted, “In the lab, ModPoKI sequences with BATF and TFAP4 have shown the potential to enhance anti-tumor activity in CAR-T cells. Next, we need to conduct further research to determine whether adding these transcription factors will make CAR-T cells more effective for human cancer patients.”
Reference
1. Blaeschke, Franziska, et al. “Modular pooled discovery of synthetic knockin sequences to program durable cell therapies.” Cell 186.19 (2023): 4216-4234.