Due to an inadequate understanding of the modes of action of therapeutic targets, inadequate preclinical models for discovery and validation in addition to the separation of the noise from real data, in vitro diagnostics (IVD) development for biomarker identification is time-consuming. Creative Biolabs has organized a staff of outstanding scientists who have engaged in IVD liquid biopsy testing for many years. We offer accurate and effective solutions for researchers who are committed to discovering protein biomarkers for IVD development.

Of all the biomolecules that can be used to diagnose and characterize cancers through liquid biopsy, protein biomarkers were among the first analytes used to assess cancers through blood tests. Protein biomarkers have been approved for assessing tumor burden in patients who have been diagnosed with cancer, particularly during therapy in patients with advanced cancer. These circulating protein biomarkers include carcinoembryonic antigen (CEA), prostate-specific antigen (PSA), beta-human chorionic gonadotropin (β-hCG), human epididymis protein 4 (HE4), alpha-fetoprotein (AFP), fibrin-degradation product (FDP), alanine aminotransferase (ALT)/aspartate aminotransferase (AST), lactate dehydrogenase (LDH), carbohydrate antigen 125 (CA 125), carbohydrate antigen 15-3 (CA 15-3), carbohydrate antigen 19-9 (CA 19-9), carbohydrate antigen 27-29 (CA 27-29). Studies have indicated that protein biomarkers may also prove valuable for the detection of early cancers.
In standard clinical protein biomarker assays, a single or a small number of prespecified tumor-associated antigens are typically targeted using immune-based methods. Although with high sensitivity and easy to be automated, there are limitations that antibodies can detect multiple proteoforms and non-specifically bind to interfering compounds generating false positives. Another technical issue affecting immunoassays is the limited dynamic range.
As an alternative to address some of these issues, high-throughput proteomics methods can go beyond detecting predetermined panels of biomarkers, complementing genomic and transcriptomic approaches in probing the molecular signature of the underlying cancer. Moreover, proteomics can provide additional information about concentrations of expressed proteins and their post-translational modifications, offering unique approaches to analyze and stratify cancer types.

As the most widely used proteomics approach, MS ionize analytes can detect and analyze ions in the gas phase. Different ionization methods, such as electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI), surface-enhanced laser desorption/ionization (SELDI), can be coupled with different analyzers to achieve different analytical characteristics. Moreover, MS instruments can be combined in tandem (MS/MS) to offer structural information on analyzed ions. Generally, MS-based proteomic methods can yield large targeted or unbiased datasets with high precision and resolution and they offer lower routine costs, higher specificity, and throughput, as well as the possibility to multiplex assays.
Microarrays are solid surfaces that immobilize up to thousands of purified or synthesized proteins at high densities. Characterized by high-throughput, high sensitivity, and robustness, protein arrays are used to quantify large predetermined panels of proteins by capturing a wide range of protein binding activities. There are several array technologies according to the captures on the solid surfaces: 1) Arrays with immobilized antibodies are a low-cost method to profile the expression of many proteins in parallel, but they rely on using pre-existing antibodies for targets of interest. 2) Functional protein arrays contain complements of purified protein and can be used for unbiased assays of the entire circulating proteome. 3) Reverse-phase lysate microarrays allow lysates from liquid biopsy to be printed on a micro-array and quantified by immunochemical methods in a massively parallel fashion. Although these arrays are typically cheaper than functional ones, they are limited by the availability of antibodies.
The aptamer-based technology is another high-throughput proteomics technology with potential use in protein biomarker discovery. This technology is a multiplex quantitative affinity-based assay where immobilized aptamers bind to proteins with high specificity and affinity.
Technologies for protein biomarker identification are being improved at a rapid rate. Multidisciplinary teams at Creative Biolabs work together closely to address these scientific and technical challenges and some of the challenges are being addressed. Creative Biolabs offers multi-analyte tests for liquid biopsy that include evaluations of circulating tumor cells (CTCs), cell-free tumor DNA (ctDNA), cell-free tumor RNA (ctRNA), exosome, tumor-educated platelets, proteins, metabolites, and nucleosome. Equipped with world-leading technology platforms, including standard clinical protein biomarker identification platform and high-throughput proteomics platforms (MS, protein array platform and aptamer-based platform), we offer high-specificity, high-throughput and cost-effectiveness protein biomarker detection services.
If you are interested in our protein biomarker detection services, please feel free to contact us for more details.
1. Liquid Biopsy-Based Monitoring of Protein Biomarker Heterogeneity in Single Circulating Therapy-Resistant Cancer Cells
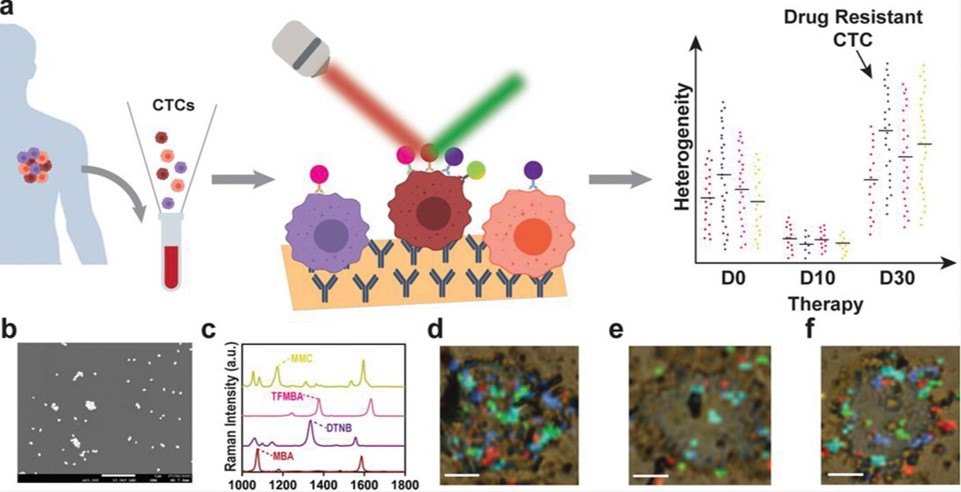 Fig.1 Schematic of CTC analysis platform.1,3
Fig.1 Schematic of CTC analysis platform.1,3
This study presented a microfluidic liquid biopsy platform for analyzing circulating tumor cells (CTCs) protein biomarker heterogeneity at the single-cell level. Using an alternating current-induced microfluidic system, individual CTCs were captured and analyzed via surface-enhanced Raman scattering spectroscopy. The platform effectively isolated single CTCs from peripheral blood mononuclear cells and detected multiple protein biomarkers on each CTC. It was shown that the platform can classify CTCs into subpopulations based on cancer-related protein changes in response to treatment. This allows for the identification of therapy-resistant CTC subpopulations, aiding clinicians in monitoring and targeting resistant cancer cells. This technology holds significant potential for disease diagnosis and treatment monitoring, advancing cancer heterogeneity research.
2. Small Extracellular Vesicle Protein Biomarkers for Pediatric Ewing Sarcoma Identified by Proteomics
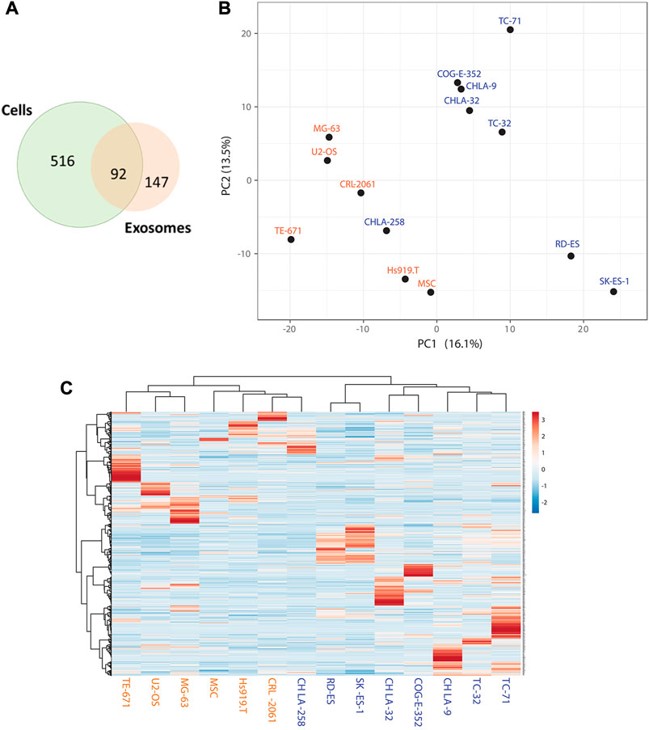 Fig.2 Identification of enriched sEV protein markers for EWS.2.3
Fig.2 Identification of enriched sEV protein markers for EWS.2.3
This research investigated Ewing Sarcoma (EWS) cell lines and their derived small extracellular vesicles (sEVs) through label-free quantitative proteomic profiling to identify protein markers enriched in EWS sEVs for validation and development as liquid biopsy markers. Researchers compared EWS cells with different EWS-ETS fusions to non-EWS and benign control cell lines. The proteomic analysis revealed common markers present in both EWS cells and their corresponding sEVs, as well as markers specifically enriched in EWS-derived sEVs. One such biomarker, UGT3A2, was identified as being highly specific to EWS cells and their derived sEVs. Clinical validation of UGT3A2 expression in tumor samples and plasma-derived sEVs confirmed its specificity to EWS, supporting its potential as an EWS liquid biopsy marker.
References
For Research Use Only.
