At the frontier of cutting-edge scientific advancements, Creative Biolabs is proud to be offering our comprehensive research-grade drug discovery service designed specifically for Klebsiella infection research. Our objective is to empower researchers by providing them with state-of-the-art research tools and technological know-how.
1. High-throughput screening.
Our high-throughput screening system employs sophisticated robotics, data processing, and control software to expeditiously perform millions of biochemical, genetic, or pharmacological assays. This allows scientists to identify active compounds, genes, or antibodies relevant to Klebsiella infection.
2. Medicinal chemistry.
Our medicinal chemistry service supports your drug discovery with lead optimization, SAR analysis, synthesis of drug precursors and more. Our team of experienced medicinal chemists provides innovative solutions for your unique research.
3. Pre-clinical testing.
We perform rigorous pre-clinical testing services to enable a smooth transition from research to clinical trials. This includes careful assessment of drug safety and dosage.
4. Pharmacokinetics & pharmacodynamics.
Investigate how potential anti-Klebsiella drugs interact within the body. Our service includes absorption, distribution, metabolism, and excretion (ADME) studies and drug-receptor interaction profiling.
5. Comprehensive analysis & reports.
We provide transparent, detailed, and easy-to-understand reports. Our experts will analyze the data and provide insights to help you make informed decisions on your drug development journey.
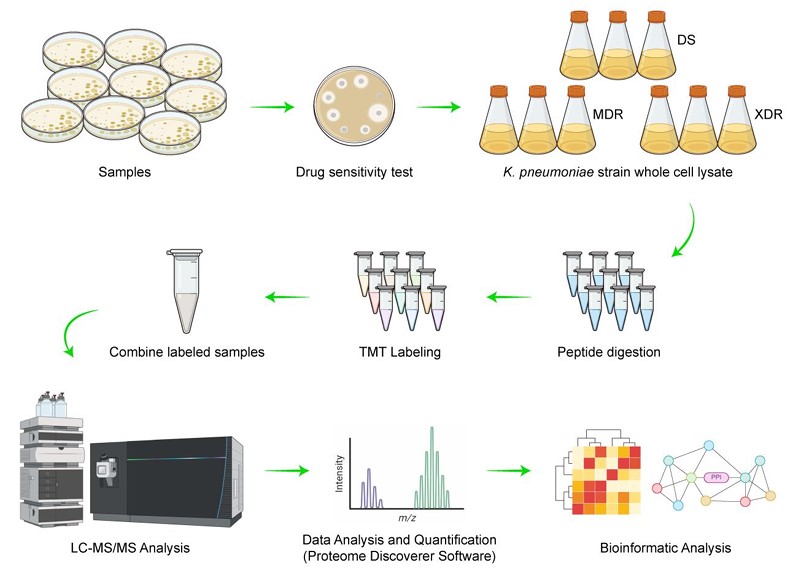 Fig. 1 Workflow of the quantitative proteomic analysis of differentially expressed proteins in Klebsiella.1
Fig. 1 Workflow of the quantitative proteomic analysis of differentially expressed proteins in Klebsiella.1
The rise in multidrug-resistant Klebsiella infections poses significant global health challenges. At Creative Biolabs, our drug discovery service specializes in Klebsiella research, aiming to pave the way for the formulation of research-grade therapeutic drugs. Please contact us to advance your Klebsiella infection research together.
Reference: