MALDI-TOF Imaging Service for Organelle Metabolic Research
Visualizing In Situ Metabolism
The interactions between a system's genome and its environment result in metabolites, representing endogenous molecules that are the end products of gene expression. To determine their biological and pharmaceutical roles, it is important to know the spatial distribution of bioactive metabolites.
Scientists at Creative Biolabs have accumulated extensive experience in organelle metabolic research. The metabolic profile of different tissue types of cancer samples can be compared using MALDI-TOF MSI. The process consists of tissue preparation, matrix application, data acquisition, data analysis, and image construction. A wide range of metabolites for the entire sample set can be covered utilizing both negative and affirmative ion detection in serial tissue sections. The MALDI TOF is highlighted through spatial and efficient detection of cancer-related analytes. It is possible to identify differential metabolites that can be further developed as diagnostic and prognostic markers for cancer.
Advantages of MALDI-TOF Imaging for the Analysis of Metabolites
-
In situ labelless molecular imaging of endogenous or exogenous metabolites.
-
Allows the identification of ionizable molecules in tissue sections or individual heterogeneous organ samples.
-
Determine the distribution of ionizable molecules present.
-
Simultaneously analyze many types of metabolites.
-
Used for pathological analysis, understanding of pharmaceutical mechanisms, and the identification of biomarkers.
Publications Sharing
Publication 1
Method Used: MALDI-MSI
Journal: Nature Metabolism
IF: 20.8
Research Findings: Combining MALDI-MSI with isotope presents an advanced approach with high spatial resolution. This approach may be used for the identification of the region's specific metabolic disturbances that are associated with lesions and throughout recovery, such as an unexpected metabolic anomaly in cells that appear normal during a recovery stage. These results may help understand the homeostatic function of the kidney microenvironment.
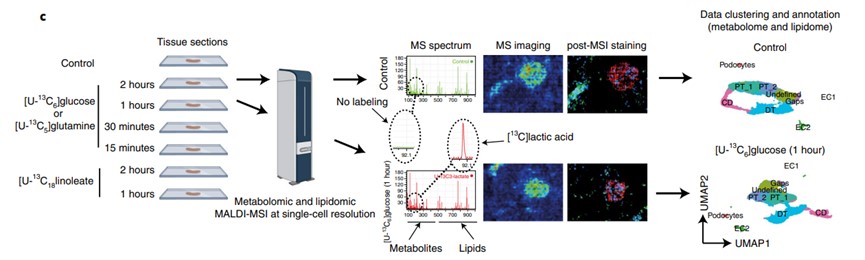 Fig.1 The metabolome and lipidome profile by MALDI-MSI.1
Fig.1 The metabolome and lipidome profile by MALDI-MSI.1
Publications Sharing
Publication 2
Method Used: Integrated MSI and LC-MS techniques
Journal: Metabolites
IF: 4.1
Research Findings: The ability to visualizein situmetabolism of endogenous metabolites or dietary phytochemicals by the most recent MALDI-MSI technology has been described in this review. Technical issues and new challenges that must be addressed for the efficient and widespread application of MSI in a variety of scientific areas, such as biological, medical, or nutritional research have been discussed by the authors.
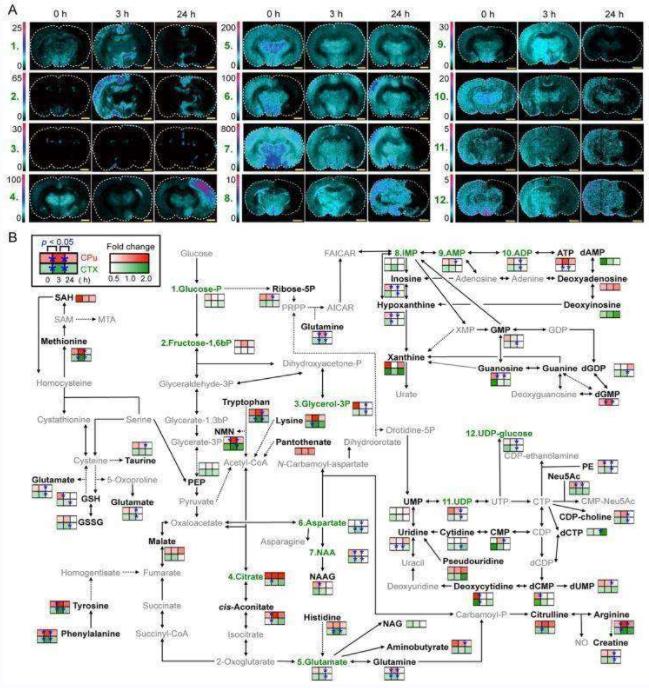 Fig.2 Integrated MSI and LC-MS techniques for visualizing spatiotemporal metabolite distribution.2
Fig.2 Integrated MSI and LC-MS techniques for visualizing spatiotemporal metabolite distribution.2
Please contact me for more information on organelle metabolic research.
References
-
Wang G, et al. Analyzing cell-type-specific dynamics of metabolism in kidney repair. Nature Metabolism. 2022 Sep;4(9):1109-18.
-
Fujimura Y, Miura D. MALDI mass spectrometry imaging for visualizing in situ metabolism of endogenous metabolites and dietary phytochemicals. Metabolites. 2014 May 5;4(2):319-46.
For Research Use Only | Not For Clinical Use


 Fig.1 The metabolome and lipidome profile by MALDI-MSI.1
Fig.1 The metabolome and lipidome profile by MALDI-MSI.1
 Fig.2 Integrated MSI and LC-MS techniques for visualizing spatiotemporal metabolite distribution.2
Fig.2 Integrated MSI and LC-MS techniques for visualizing spatiotemporal metabolite distribution.2
 Download our brochure
Download our brochure