Gene Insertion
The gene-editing technology opens up a paradigm of the possibility of precise modifications and manipulation of genome sequences of interest to achieve a therapeutic effect by enabling targeted disruption, insertion, excision, and correction in both ex vivo and in vivo settings. Gene editing-based therapeutics have a unique ability to elaborately and controllably modify the genome, including the correction of mutations that cause disease, the addition of therapeutic genes to specific sites in the genome, and the removal of deleterious genes or genome sequences.
Gene Insertion
Traditional gene therapy including plasmid DNA or integrating viral vectors can be successfully used to insert a gene of interest into the genome; however, the insertion site of these viruses is an inability to control, which results in the possibility of insertional mutagenesis and its regulatory sequence can be possibly incorporated at undesired sites. Custom-designed and engineered nucleases enable precise and targeted modification of the genome, and the targeted therapeutic genes are introduced into pre-determined sites in the genome, such as genomic "safe harbor" which could alleviate risks of insertional mutagenesis and enhance the efficiency in a site-specific manner to enable high levels of gene expression.
DNA-binding modules are coupled to nuclease and introduce double-strand breaks (DSBs) at the chosen site. Once the nucleases-induced DSBs have been made, DSBs are then normal repaired to insert a sequence of choice by either non-nonhomologous end joining (NHEJ) or homology-directed repair (HDR) depending on the cell state and the presence of a repair template. Gene insertion can be exploited to introduce targeted genome alterations in a wide range of organisms and cell types.
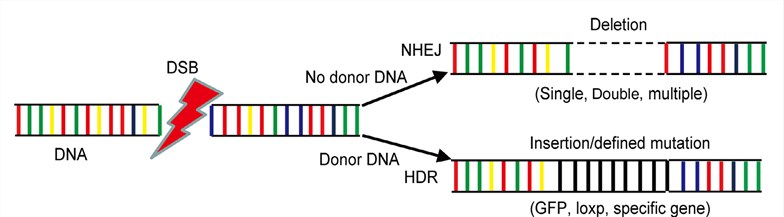 Figure 1. The mechanisms of gene editing. (Li, 2017)
Figure 1. The mechanisms of gene editing. (Li, 2017)
HDR-mediated Insertion
HDR is a repair mechanism reliant upon homologous recombination (HR) that using homologous donor DNA as templates with homology arms which flanked by DNA segments homologous to the targeted sequence; it allows introducing targeted insertion of a correct DNA sequence at a specific genomic site. In this way, gene editing nuclease-mediated HDR can be exploited to introduce precise alterations or efficiently exogenous DNA insertions of up to 7.6kb at or near the target site of the break.
NHEJ-mediated Insertion
An alternative repair mechanism for targeted transgene insertion is to use nuclease-induced DSBs to create defined overhangs on the donor DNA and the endogenous site, leading to NHEJ-mediated ligation of the directly insert variable-length DNA at the site of the break without using a homologous template. NHEJ pathway can rapidly and efficiently ligate the two broken ends; however, it is also an error-prone process in which the genetic information is occasionally lost leading to the loss of encoding protein functions. NHEJ-mediated insertion is often used to introduce small disruptive insertions and/or deletions at the break site and can be exploited to disrupt a target gene.
Such gene insertion strategies can link DNA sequences that encode reporter genes or peptide tags to the corresponding endogenous locus and provide better monitor over transgene copy number and expression levels under regulatory elements. The therapeutic transgene may also be inserted into a new locus to restore missing gene function, including identified "safe harbor" loci. Up to now, gene insertion allows being applied to various areas of gene and cell therapy to achieve the therapeutic effect, such as stably conferring novel functions against different diseases or the protection against repeat infection.
References
- Kim, H.; Kim, J. S. (2014). A guide to genome engineering with programmable nucleases. Nature Reviews Genetics. 15(5): 321-334.
- Li, M.; Wang, D. (2017). Gene editing nuclease and its application in tilapia. Science Bulletin. 62(3): 165-173.
