Mechanisms of Gene Editing
An overview
Gene editing has the ability to make highly specific changes in the DNA sequence of a living organism, essentially customizing its genetic makeup. This process is performed by using enzymes, particularly nucleases that have been engineered to target a specific DNA sequence, where they introduce cuts into the DNA strands, enabling the removal of existing DNA and the insertion of replacement DNA.
Mechanisms of Gene Editing
It is an important discovery in the field of gene editing that targeted DNA double-strand breaks (DSBs) could be used to stimulate the endogenous cellular repair machinery. Breaks among the DNA are typically repaired via one of two major pathways: homology-directed repair (HDR) or nonhomologous end-joining (NHEJ).
HDR is based on strand invasion of the broken end into a homologous sequence and subsequent repair of the break in a template-dependent manner. The functions of NHEJ to repair DSBs without a template through direct re-ligation of the cleaved ends. There are four approaches for gene editing--gene knockout (KO)/mutation, gene deletion, gene correction, and gene insertion.
 Figure 1. Mechanisms of double-strand break repair. (Maeder, 2016)
Figure 1. Mechanisms of double-strand break repair. (Maeder, 2016)
Gene knockout is the simplest form of gene editing and utilizes the error-prone nature of NHEJ to introduce small indels at the target site. Classical NHEJ directly re-ligates unprocessed DNA ends. The other alternative-NHEJ, also known as microhomology-mediated end joining, or MMEJ, requires end-resection followed by annealing of short single-stranded regions of microhomology and subsequent DNA end ligation.
 Figure 2. Gene knockout. (Gaj, 2016)
Figure 2. Gene knockout. (Gaj, 2016)
In addition to the relatively minor indels resulting from NHEJ, it is possible to delete large segments of DNA by flanking the sequence with two DSBs. Indeed, it has been shown that simultaneous introduction of two targeted breaks can give rise to genomic deletions up to several megabases in size. This approach is crucial within therapeutic strategies that may require the removal of an entire genomic element. For example, an enhancer region, has been proposed for the treatment of hemoglobinopathies by deletion of the BCL11A erythroid-specific enhancer region.
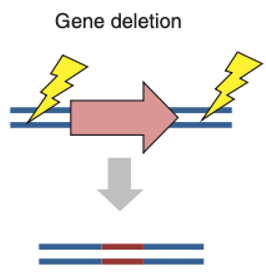 Figure 3. Gene deletion. (Gaj, 2016)
Figure 3. Gene deletion. (Gaj, 2016)
Due to opposed to the unpredictable mutations resulting from NHEJ, targeted DSBs can induce precise gene editing by stimulating HDR with an exogenously supplied donor template. Active mainly during the S and G2 phases of the cell cycle, HDR naturally utilizes the sister chromatid as a template for DNA repair. Any sequence differences present in the donor template can thus be incorporated into the endogenous locus to correct disease-causing mutations, which has been demonstrated in many proof-of-concept studies.
 Figure 4. Gene correction. (Gaj, 2016)
Figure 4. Gene correction. (Gaj, 2016)
Gene insertion is the kind way that the addition of one or more genes into a DNA sequence. This technique is common to be performed with plasmid DNA or integrating viral vectors. However, in the conventional gene insertion method, the insertion site cannot be controlled. An alternative mechanism for targeted transgene insertion is to employ nuclease-induced DSBs to create compatible overhangs on the donor DNA and the endogenous site, leading to NHEJ-mediated ligation of the insert DNA sequence directly into the target locus.
References
- Maeder, M. L.; Gersbach, C. A. (2016). Genome-editing technologies for gene and cell therapy. Molecular Therapy. 24(3): 430-446.
- Gaj, T.; et al. (2016). Genome-editing technologies: principles and applications. Cold Spring Harbor perspectives in biology. 8(12), a023754.
