Creative Biolabs is pleased to introduce an unparalleled de novo single domain antibody sequencing service that has been developed, based on our world-leading platform of de novo antibody sequencing using our proprietary Database Assisted Shotgun Sequencing (DASS) technology. Soluble and functional single domain antibodies can be sequenced by subunit with 100% coverage and dozens of successful cases proving 100% accuracy.
Single domain antibody (sdAb) is a kind of antibody fragment consisting of a monomeric antibody variable domain (VH) and lacking the light chain and CH domain of the heavy chain in the conventional Fab region of an IgG. The first sdAb was derived from the VHH domain of heavy-chain antibodies (HCAbs) identified in camelids (llama, alpaca, camel, dromedary, etc.). The VNAR domain of cartilaginous fish (e.g., shark) antibodies (known as IgNAR, immunoglobulin new antigen receptor) is another qualified source for sdAb development. An alternative approach is to split the dimeric variable domains from normal IgGs in humans or mice into monomers by camelizing a few critical residues.
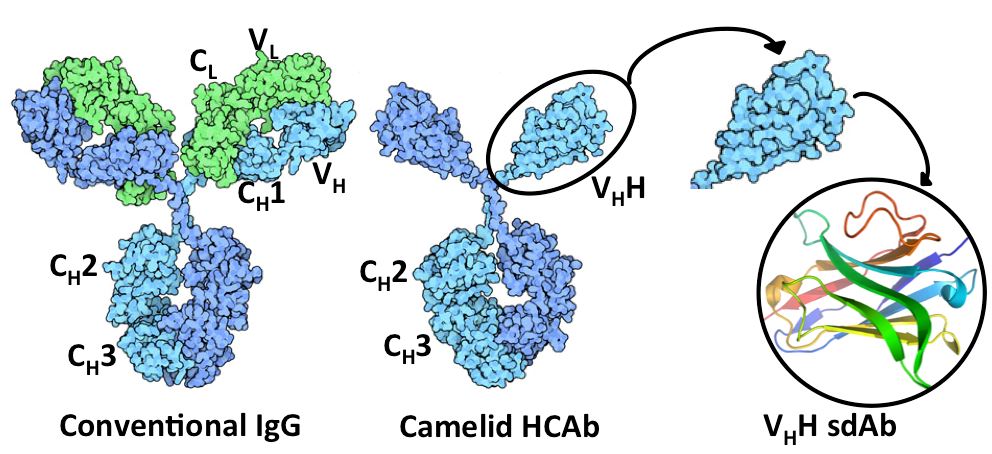 Fig. 1 Structural comparison between conventional IgG and camelid HCAb.
Fig. 1 Structural comparison between conventional IgG and camelid HCAb.
Similar to conventional antibodies, sdAbs are able to bind selectively to a specific antigen. sdAbs can be easily produced in bacteria or yeast in large quantities with high stability, as well as antigen binding affinities and specificities. With their size in the nanometer range, sdAbs take great advantage of stability and penetrability, which are essential for the development of antibody-based drugs or diagnostic tools.
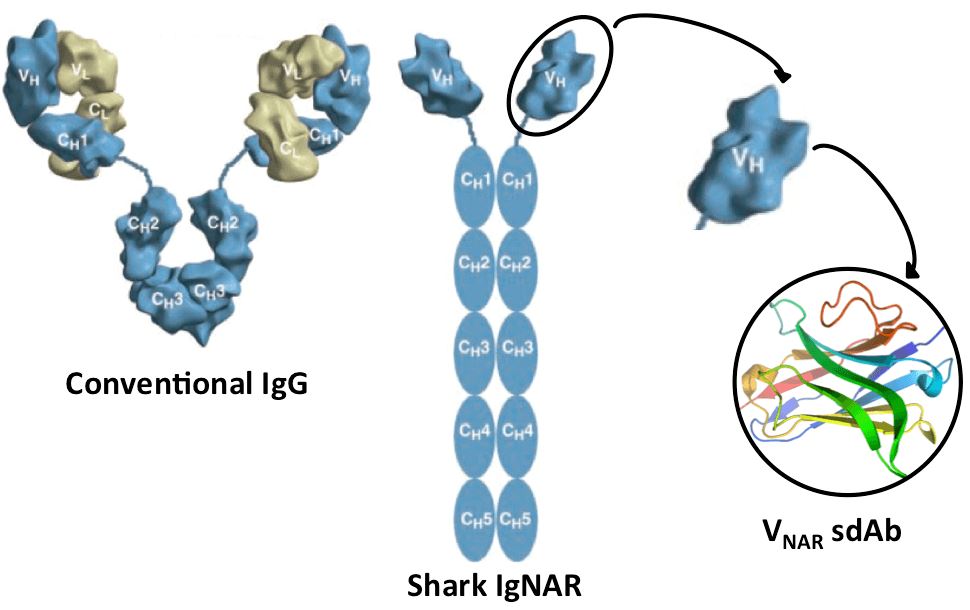 Fig. 2 Structural comparison between conventional IgG and shark IgNAR. (Robyn L Stanfield, 2004)
Fig. 2 Structural comparison between conventional IgG and shark IgNAR. (Robyn L Stanfield, 2004)
With the most advanced DASS technology, Creative Biolabs provides services helping customers obtain the sequence information of sdAbs, which plays key roles in the development of therapeutic or diagnostic sdAbs. Our world-leading de novo antibody sequencing platform can achieve successful sequencing of sdAbs with diverse origins with 100% coverage and 100% accuracy. For instance, the full length of the camelid heavy chain antibody (HCAb) sequence could be obtained by mass spectrometric de novo sequencing. Leu/Ile ambiguities could be resolved by homology comparisons with known VHH sequences. Moreover, sdAb conjugates or immobilized sdAbs can be successfully sequenced through our DASS technology as well.
Please find more general features of our DASS technology in de novo antibody sequencing service, or directly inquire us for more details.
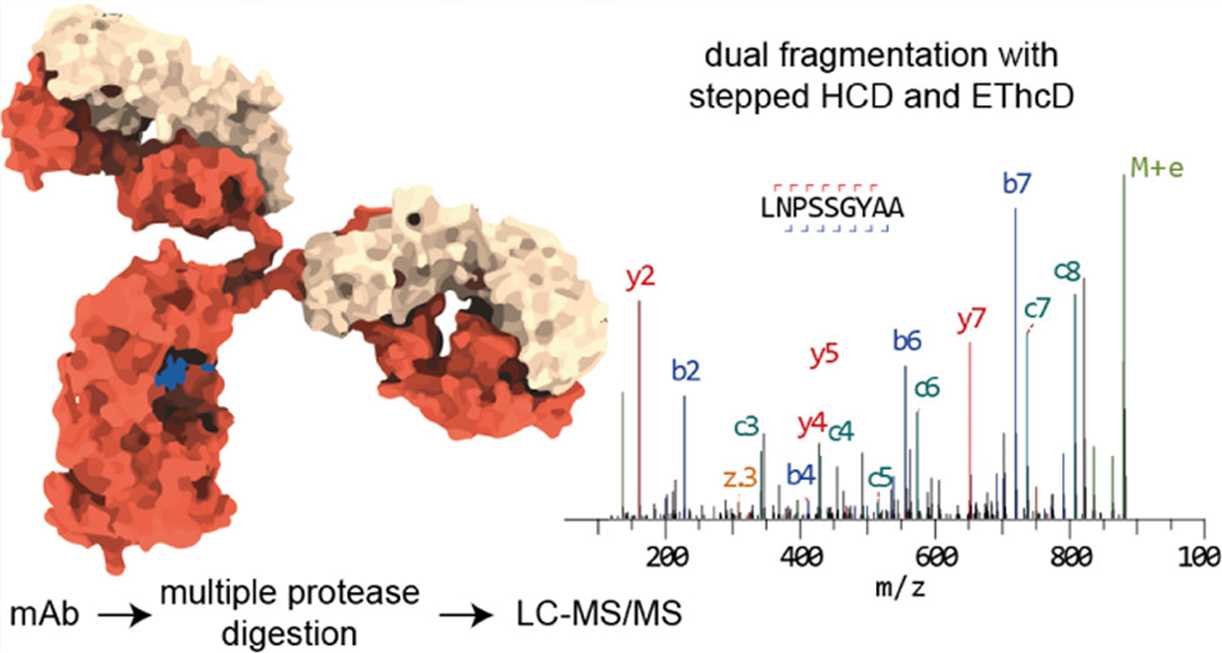 Fig. 3 De novo protein sequencing of monoclonal antibodies. (Weiwei Peng, 2021)
Fig. 3 De novo protein sequencing of monoclonal antibodies. (Weiwei Peng, 2021)
Antibody sequence information is very important for understanding the structural basis of antigen binding. In this article, the researchers demonstrated a method of direct de novo sequencing of monoclonal IgG from purified antibodies. This method uses a group of complementary proteases to cleave antibodies to produce suitable peptides for bottom-up de novo sequencing by liquid chromatography-tandem mass spectrometry (LC-MS/MS). This method can cover the whole sequence of anti-HER2 monoclonal antibodies, and the accuracy of the variable region is 99%. They used this method to sequence the widely used anti-FLAG-M2 mouse monoclonal antibody and successfully verified the antibody by reconstructing the high-resolution crystal structure of Fab and proving its binding to the FLAG-labeled target protein in Western blotting analysis.
De novo sequencing of single domain antibodies is a method that does not depend on known DNA sequences and is used to determine the amino acid sequence of antibody proteins. This technique is often used for antibodies that do not have available genetic information, such as those obtained from natural sources or through non-traditional methods. De novo sequencing involves the use of mass spectrometry to analyze the peptides of antibody proteins and to infer the complete amino acid sequence by reconstructing the continuity between the peptides.
The first is the difficulty of identifying the overlapping regions of peptides and the complexity of sequence analysis. Mass spectrometry data may become complex due to factors such as peptide size, charge, and hydrophilicity, which require highly accurate instruments and advanced data analysis techniques to accurately identify and sort. In addition, antibodies may have glycosylation or other post-translational modifications, which will affect the results of mass spectrometry and increase the difficulty of de novo sequencing.
First of all, the antibody samples need to be purified to ensure the accuracy of mass spectrometry analysis. Then, the antibody protein was hydrolyzed by enzymes and cut into smaller peptides. These peptides are then analyzed by a mass spectrometer, which measures the mass and charge of each peptide to generate a mass spectrum. Using these data, researchers use special software to identify and sequence peptides and reconstruct the original protein sequence. This process requires repeated verification and alignment to ensure the accuracy of the sequence.
First of all, the use of the high-resolution mass spectrometer can greatly improve the resolution and accuracy of peptides, so that peptides in complex samples can be identified more accurately. Secondly, optimizing the enzymatic hydrolysis steps can improve the coverage and uniformity of the peptide map, which is very important for the complete reconstruction of the sequence. Digestion with a variety of enzymes can produce peptides at different cutting points and increase overlapping regions, thus improving the accuracy of sequence reconstruction. Finally, the use of advanced data processing and analysis software can process mass spectrometry data more effectively, identify and correct potential errors, and ensure the accuracy of sequence analysis.
The main difference between de novo sequencing of single domain antibodies and traditional DNA sequencing lies in their analytical objects and methods. DNA sequencing is to directly read the gene sequence of the antibody and obtain the amino acid sequence encoded by the gene sequence. This method relies on available genetic information and is usually used for sequence analysis of known genomes or cloned antibodies. In contrast, de novo sequencing is a protein analysis method, starting directly from the antibody protein itself and using mass spectrometry to analyze the digested peptides and reconstruct the amino acid sequence of the protein. This method does not rely on DNA information and is suitable for antibodies without genetic information or difficult to obtain DNA sequences, especially in the study of antibodies found from natural sources or complex biological samples.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
