Ribosome display technology is a novel in vitro technology for screening and molecular evolution of biological libraries. Ribosome display is performed in a cell-free system in vitro, without involving living cells or phages, thus avoiding the limitations of transformation efficiency necessary to import the original library into cells, and the library capacity can reach 1015. Therefore, high-affinity antibodies can be screened against a wide range of target antigens up to the pM level. In addition, ribosome display technology is also commonly used in the affinity maturation process of antibodies, which can increase the affinity of antibodies by more than 30-fold, to 10–12 mol/L. Based on our rich field experience and ribosome display platform, Creative Biolabs provides comprehensive services to support the development and application of eukaryotic ribosome display systems.
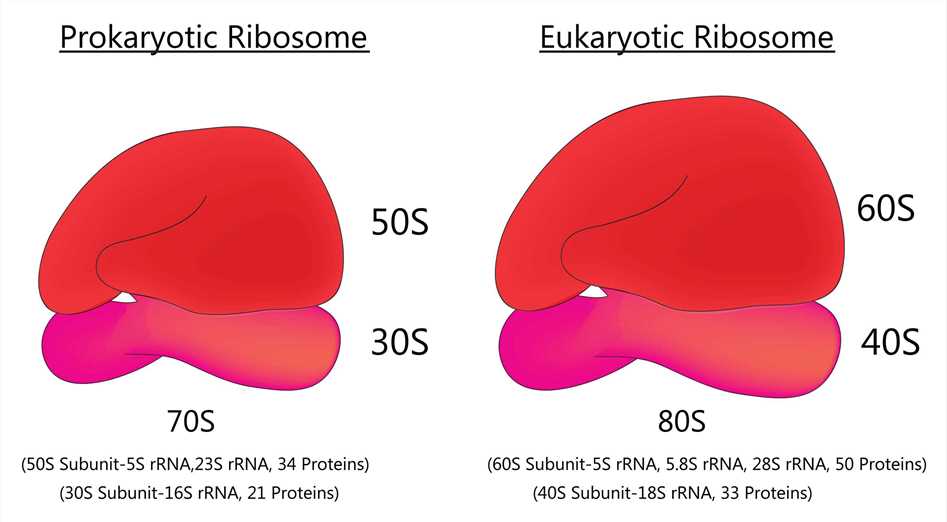 Fig 1. Prokaryotic ribosome and Eukaryotic ribosome
Fig 1. Prokaryotic ribosome and Eukaryotic ribosome
About 20 years ago, in vitro display techniques such as phage display, bacterial display, and yeast display were first used for antibody screening. To date, many antibodies have been screened and improved using these methods. Although still not widely used, many of the antibodies screened by in vitro display techniques are difficult, if not impossible, to obtain by immunizing animals. Ribosome display is a cell-free system for in vitro selection of proteins and peptides from large libraries. It uses the principle of coupling individual nascent proteins (phenotypes) to their corresponding mRNAs (genotypes) through the formation of stable protein-ribosome-mRNA complexes (ARM). This allows for the simultaneous isolation of functional de novo proteins by affinity for the ligand as well as the encoding mRNA, which can then be transformed and amplified into DNA for further manipulation, including repetitive cycling or protein expression. Ribosome display has many advantages over cell-based systems such as phage display; in particular, it can display very large libraries without the constraints of bacterial transformation. It is also suitable for generating toxic, proteolytically hydrolysis-sensitive, and unstable proteins and allows for the incorporation of modified amino acids at specific sites.
A study reports on the eukaryotic ribosome display system, also known as the ARM display system. This technique was used to select functional single-chain antibody fragments in a coupled rabbit reticulin lysate system. Deletion of stop codons from the PCR fragments was again used to generate eukaryotic ARM complexes in a simple and rapid procedure. ARM complexes were generated by cell-free expression of PCR fragments followed by specific capture on antigen-coated magnetic beads. DNA was recovered from the captured mRNA by amplification by a novel in situ RT-PCR procedure without dissociating the ribosome complex. Starting from an anti-progesterone hybridoma cell line (DB3), the ARM complex, consisting of a three-domain antibody fragment (Vн/K) retained the detailed specificity of the monoclonal antibody to DB3, which implies that the nascent protein is folding correctly. The specific progesterone-binding complex can be enriched from mutant libraries by antigen selection, which can be increased approximately 104-fold in one display cycle. An ARM display modification has been described in which oxidized/reduced glutathione and q β RNA-dependent RNA polymerase are included in the translation mixture to display folded proteins and introduce mutations in mRNA to promote protein evolution in vitro. Eukaryotic display systems have also been used to select an enzyme, sialyltransferase ii, from single-well cDNA libraries in microplates.
Creative Biolabs has a wealth of knowledge and experience in ribosome display. We would be happy to share with you our knowledge and experience related to the eukaryotic ribosome display system.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.