Ribosome display systems have been reported as early as about 30 years ago, but due to the limitations of the method, they have not been widely used. Nowadays, the ribosome display technology has been improved, and the technology has been more widely used. In addition to biological research, ribosome display technology will be more widely used in chemical biology and materials chemistry. For example, peptide aptamer screening is performed from a library of conductive polymers composed of hydrophilic amino acids, and the bound aptamers are measured by quartz crystal microbalance, and their secondary structures are analyzed by circular dichroism. Based on our rich field experience and ribosome display platform, Creative Biolabs provides comprehensive services to support the development and application of prokaryotic ribosome display systems.
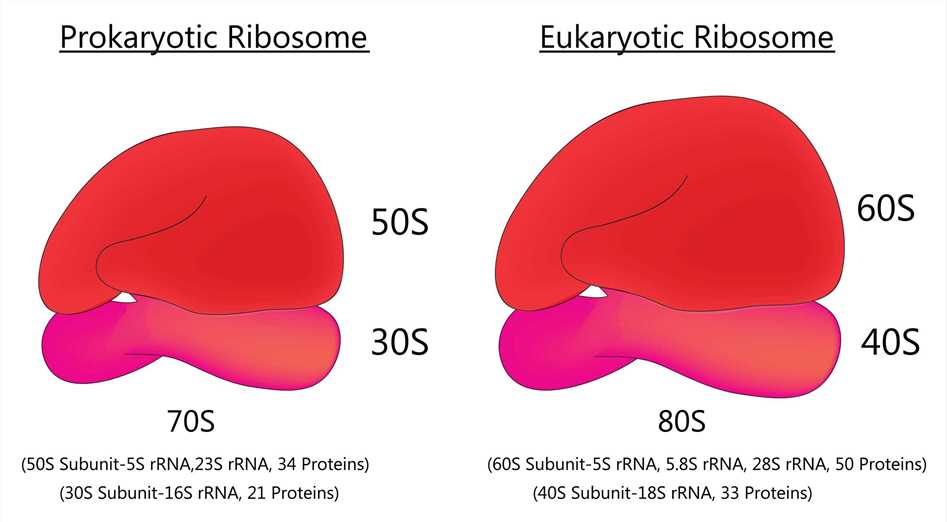 Fig 1. Prokaryotic ribosome and Eukaryotic ribosome
Fig 1. Prokaryotic ribosome and Eukaryotic ribosome
Ribosome display technology has been around for some time, but due to various limitations, this technology has not been as widely used as phage display technology. The main reason for this is that it is difficult to prepare stable trimeric complexes in cell extracts, and cell-free protein synthesis systems contain many inhibitory factors that inhibit effective ribosome display. With further research and technological development, the ribosome display technique has now been refined and updated to provide a more stable performance.
Ribosome display technology is performed in vitro and avoids some of the limitations of cell-based display systems. By forming mRNA-ribosome-protein trimers in a cell-free system, nascent proteins are associated with their associated mRNA molecules, and the genetic material is selected by the functional properties of the proteins. In recent years, researchers have utilized ribosome display technology for extensive studies and have improved the traditional ribosome display technology to successfully screen a wide range of macromolecules, such as antibodies, enzymes, peptides, and some small molecules that have a specific affinity for target molecules.
About 30 years ago, scholars constructed a very large peptide library using an in vitro protein synthesis system. The first published description of ribosome display is based on a prokaryotic ribosome display system. Specifically, it utilizes the coupled E. coli S30 system for peptide selection, called "polysome display". Synthetic DNA libraries encode random peptide sequences, and translation is stopped by the addition of chloramphenicol to produce the polysome complex. Specific complexes displaying interacting peptide epitopes are captured by immobilized antibodies through microsieve wells. The captured polysomes were disrupted with EDTA to release bound mRNA, which was then transformed and amplified into cDNA by RT-PCR. This procedure was subsequently modified to reveal folded single-chain antibody fragments. As research progressed, improved prokaryotic ribosome display techniques emerged. The modified prokaryotic ribosome display technique generates mRNA-ribosome-protein trimers by deleting the 3' end termination codon of DNA. The translation mixture includes some additional components such as protein disulfide bond isomerase, vanadyl ribonucleoside complex, and anti-ssrA antisense oligonucleotide (which prevents the addition of the carboxyl terminus of the 11-amino acid peptide encoded by the ssrA gene) for facilitating the folding of the antibody fragments, stabilizing the mRNAs, and inhibiting the action of the ssrA RNAs, respectively. To avoid the disruptive effect of dithiothreitol on the folding of the antibody structural domain by reducing the disulfide bond, scholars transcribed and translated the mRNAs separately and introduced the mRNAs into the uncoupled E. coli S30 translational system, which lacks DTT.
Creative Biolabs has a wealth of knowledge and experience in ribosome display. We would be happy to share with you our knowledge and experience related to prokaryotic ribosome display.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.