The scientific study describes a cutting-edge methodology that uses yeast surface display (YSD), next-generation sequencing (NGS), and artificial intelligence/machine learning (AI/ML) to quickly identify de novo humanized single domain antibodies
(sdAbs) with optimized VHH specificity and commendable early-stage development profiles. It shows how NGS can analyze large sequence spaces and how AI/ML can be used to design new sequences with better potency or developability. The ability of long
short-term memory (LSTM) networks, a subset of recurrent neural networks, to capture intricate correlations between amino acids that dictate the structure and function of proteins is especially noteworthy. The high-affinity binding and favorable biophysical
characteristics of the discovered VHHs are validated experimentally. Accurate predictions in silico are crucial for efficient sequence selection and enhanced developability profiles, thus supporting the use of AI/ML for specificity optimization.
By predicting which sequences are less likely to aggregate, researchers can prioritize candidates that are more likely to maintain their structural integrity and specificity in biological systems. Besides, understanding and optimizing the biophysical
properties, such as hydrophobicity and aggregation propensity, contribute to the overall developability of an antibody, affecting its manufacturability, stability, and pharmacokinetics.
1. Why is the specificity of VHH antibodies critical in research and therapeutic applications?
Specificity is vital because it determines the ability of a VHH to bind exclusively to its target antigen without cross-reacting with other molecules. High specificity ensures accurate detection, quantification, and targeted treatment of diseases, thereby
reducing off-target effects and false positives in research.
2. How does the natural diversity of camelid VHH repertoires affect specificity optimization?
The natural diversity of camelid VHH repertoires generates a wide range of distinct binding locations and frameworks. This diversity enables the selection of VHHs with high specificity and affinity for different antigens, which may then be fine-tuned
using techniques such as phage display and directed evolution.
3. What role do complementarity-determining regions (CDRs) play in the specificity of VHHs?
CDRs are hypervariable sections of the VHH that interact directly with the antigen, determining the specificity and affinity of antibody binding. Optimization frequently focuses on the CDRs modifications to improve binding characteristics while retaining
or increasing specificity.
4. Can you explain the importance of structural stability in VHH specificity optimization?
Structural stability is critical because it guarantees that the VHH retains its conformation and binding activity under a variety of circumstances. Enhanced stability helps to improve binding specificity by preserving the precise conformation required
for specific antigen engagement, even under demanding conditions like high temperatures or fluctuating pH levels.
5. What are some challenges in optimizing VHH specificity, and how can they be addressed?
There are several challenges in VHH specificity optimization, including cross-reactivity, poor affinity, and structural constraints. The unwanted binding to non-target molecules may occur as cross-reactivity, but this can be solved by improving the selection
procedure to eliminate non-specific binders. Poor affinity is often increased via affinity maturation procedures. Due to some structural constraints, it is difficult to modify the VHH without disrupting its general structure and normal functions.
However, this obstacle can be handled with the help of advanced bioinformatics and structural biology technologies.














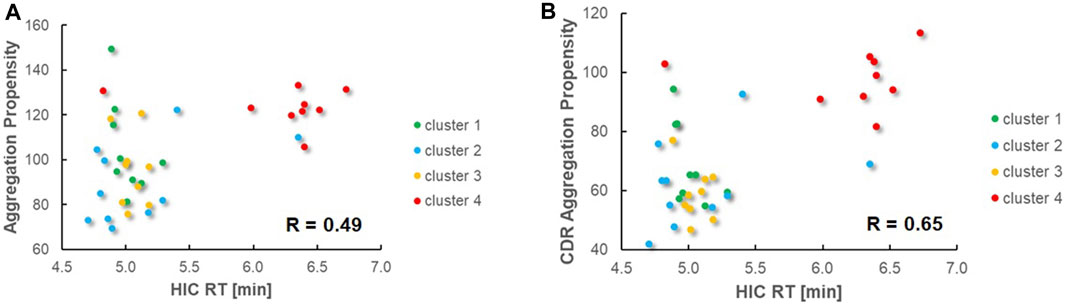 Fig. 1 Prediction and Experimental Validation of Aggregation Tendencies in VHH Domains via HIC Analysis.1
Fig. 1 Prediction and Experimental Validation of Aggregation Tendencies in VHH Domains via HIC Analysis.1















