
Whole Transcriptome Sequencing (WTS) based Bioinformatic Analysis Service
Whole Transcriptome Sequencing (WTS)
- Overview
Whole Transcriptome Sequencing (WTS) is an advanced technique that leverages next-generation sequencing (NGS) to analyze all RNA molecules in a given sample, including messenger RNA (mRNA), non-coding RNA, and other RNA species. This method offers a comprehensive snapshot of gene expression, allowing researchers to quantify all expressed genes in a cell or tissue.
What sets WTS apart is its ability to provide more than just gene expression levels. It can identify alternative splicing events, detect gene fusions, uncover RNA editing, and even reveal post-transcriptional modifications. Importantly, it also enables the discovery of novel transcripts that might have been overlooked in previous analyses. Compared to traditional methods like microarrays, which rely on pre-selected probes, WTS offers an unbiased approach that captures the full complexity of the transcriptome, even picking up on rare and low-abundance transcripts. WTS has found widespread applications in diverse fields of research. In cancer studies, it is often used to identify unique gene expression patterns in tumors, which can point to potential biomarkers or therapeutic targets.
The amount of data generated by WTS can be immense, and analyzing it often requires sophisticated bioinformatics tools. Once processed, this data provides valuable insights into gene regulation, cellular pathways, and the molecular underpinnings of diseases. Such information is crucial for understanding biological processes and is increasingly being used to guide the development of precision medicine.
In essence, Whole Transcriptome Sequencing offers researchers a powerful tool to delve deeply into gene expression and regulation. Its ability to capture the full scope of the transcriptome makes it an essential resource in fields ranging from basic biology to clinical research, significantly advancing our understanding of complex biological systems and diseases.
- Workflow
Whole Transcriptome Sequencing (WTS) for cancer research involves several key steps
- Sample Collection and Preparation: RNA is extracted from cancer tissues or cells.
- Library Preparation: RNA is enriched or depleted, converted to cDNA, fragmented, and sequencing adapters are added.
- Sequencing: High-throughput sequencing platforms generate millions of reads representing the transcriptome.
- Bioinformatics Analysis: Data filtration, alignment, differential expression analysis, variant and fusion detection, other specific analyses.
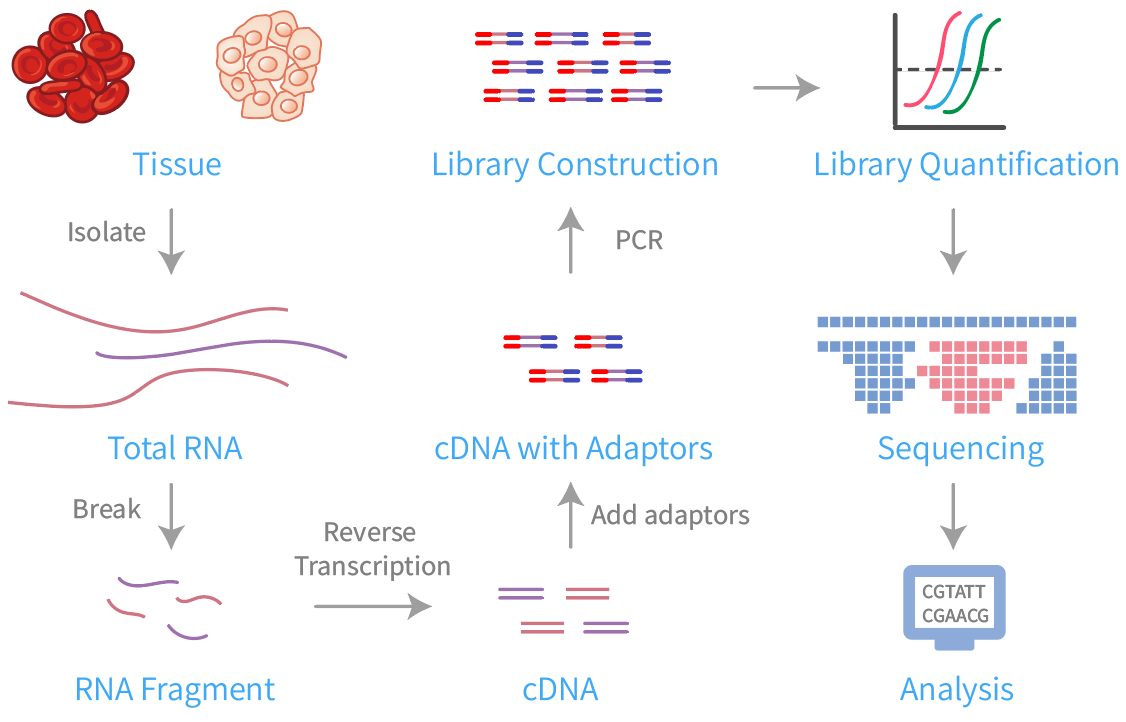 Fig.1 WTS workflow.
Fig.1 WTS workflow.
Cancer WTS Bioinformatics Analysis at Creative Biolabs
- Services
WTS data analysis plays a crucial role in cancer research by transforming raw sequencing data into actionable biological insights. This bioinformatics-driven process is essential for several key tasks, including the quantification of gene expression, differential expression analysis, and the identification of alternative splicing events, gene fusions, and mutations. In cancer, WTS data analysis enables researchers to compare tumor and normal tissue gene expression, uncovering potential oncogenes, tumor suppressors, and therapeutic targets. It also allows for the discovery of novel transcripts, such as non-coding RNAs, which may have a role in cancer progression. Furthermore, bioinformatics tools are indispensable for pathway enrichment analysis, helping to identify disrupted molecular pathways that drive cancer. The ability to visualize complex data patterns through clustering and other visualization techniques aids in identifying cancer subtypes and predicting patient outcomes. Ultimately, WTS data analysis provides the foundation for biomarker discovery and personalized treatment strategies, making it an essential component of modern cancer genomics and precision medicine.
Scientists of Creative Biolabs have developed a range of different tools for the WTS bioinformatics analysis pipeline. We can offer a series of solutions, from differential expression analysis to the identification of mutations and fusion transcripts.
- Applications
Potential applications of our WTS bioinformatics analysis encompass
- Drug Development
- Biomarker Discovery
- Evolutionary Study
- Resistance Study
- Therapeutic And Prophylactic Intervention
- Key Notes for WES Bioinformatic Analysis
The following section outlines key considerations and best practices for ensuring accuracy and reliability in WES bioinformatic analysis:
1. Quality Control of Data
Ensuring data quality is essential to prevent errors in downstream analyses. Tools like FastQC are crucial for assessing raw data quality.
2. Accurate Read Alignment
Proper alignment to the reference genome or transcriptome using tools like HISAT2 or STAR is key to ensuring correct gene expression estimates and transcript detection.
3. Normalization Methods
Applying appropriate normalization methods (e.g., TPM, FPKM) is necessary to account for differences in sequencing depth and technical biases, ensuring accurate expression data.
4. Differential Expression Analysis
Robust statistical methods (e.g., DESeq2, edgeR) are required to accurately identify significant gene expression changes between conditions, with careful attention to replicates and thresholds.
5. Data Visualization
Effective visualization tools (e.g., heatmaps, PCA) are critical for interpreting complex data, helping to identify patterns and biological insights.
Key Features and Advantages
- Compare variants in DNA and RNA.
- Identify candidate variants and genes from a tumor-normal pair.
- Annotate variants.
- Identify and annotate differentially expressed genes.
- Highly professional Ph.D. level scientists.
- Fast turnover time.
- Competitive and affordable price.
Equipped with enriched experience, profound expertise, and a state-of-the-art technic platform, Creative Biolabs is fully competent and dedicated to serving as your one-stop-solution for transcriptome sequencing (mRNAseq, lncRNAseq, circRNAseq, miRNAseq) service and corresponding bioinformatics analysis service. We can offer high-quality customized services to meet even the most specific requirements.
Please contact us for more information and a detailed quote.
Q&As
Resources
Infographics
Podcast
- WGS Bioinformatic Analysis
- WES Bioinformatic Analysis
- Targeted Sequencing Bioinformatic Analysis
- SuPrecision™ Pipeline




