Therapeutic protein fold recognition has attracted increasing attention because it is critical for studies of the 3D structures of proteins and drug design. Predicting the 3D fold of a protein is a key problem in molecular biology. Creative Biolabs has focused on the development of protein engineering services for many years and has established excellent protein engineering platforms for drug development. We provide a variety of protein fold recognition services to meet the diverse needs of our customers.
Knowledge of protein folding has an important effect on understanding the molecular function of proteins, further facilitating drug design. Computational recognition of protein folds has been a recent research hotspot in bioinformatics and computational biology. Computational prediction methods can automatically, quickly and accurately classify unknown protein sequences into specific folding categories. Many computational efforts have been made, generating a variety of computational prediction methods. In silico-based methods can be further categorized into two classes according to the learning algorithms used in prediction models: single classifier-based methods and ensemble classifier-based methods.
Single classifier-based methods use a single specific learning algorithm to build prediction models, whereas ensemble classifier-based methods use an ensemble of multiple, either similar or different, learning algorithms to build prediction models. Most of the single classifier methods used in protein fold recognition are based on support vector machines (SVM) classifier because SVM, being a well-known classification algorithm, has been highly efficient in several fields of bioinformatics, such as in protein remote homology detection, protein structural classification, and DNA-binding protein prediction.
There are three general types of ensemble classifier models. For a given n different single basic classifiers, the first type of ensemble classifier-based method uses one specific feature descriptor to encode query proteins with feature representations. For a given n different single basic classifiers and n different feature descriptors, the second type of ensemble classifier-based method uses n feature descriptors to encode query proteins with n different feature representations. For a given specific classifier and n different feature descriptors, the third type of ensemble classifier-based method uses n feature descriptors to encode query proteins with n different feature representations.
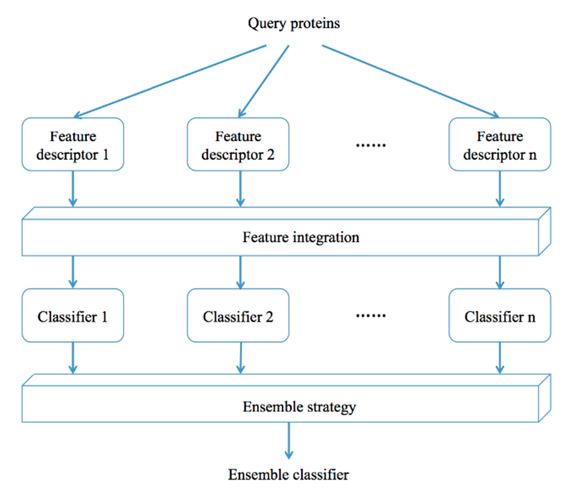 Fig.1 The construction type of ensemble-classifier models.
Fig.1 The construction type of ensemble-classifier models.
In silico-based methods can be successfully applied in protein fold recognition. In the future, these computational methods will be extensively applied in other similar but unexplored fields, such as disease-causing amino acid change prediction, protein-protein binding site, and protein-protein interaction prediction. Therefore, we can provide various protein fold recognition services, including single classifier-based methods and ensemble classifier-based methods, to meet customers' specific requirements.
Creative Biolabs has been involved in the field of protein engineering for many years and we are committed to completing your project with high quality. We have accumulated a wealth of scientific experience from our completed projects and provide you with the best services to ensure your requirements are met. If you are interested in our services, please contact us for more details.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.