Stability and affinity of proteins are important factors affecting protein function. Therefore, the selection and concomitant affinity maturation of proteins to user-defined target molecules has become a key technology for biochemical research, diagnosis, and therapy. Ribosome display technology, on the other hand, is the most effective tool for evolving proteins. Ribosome display technology is performed in vitro in cell-free media and is based on the formation of ternary ribonucleoprotein complexes between RNA, ribosomes, and proteins. Ribosome display has become one of the most widely used presentation methods today. Based on our rich field experience and ribosome display platform, Creative Biolabs provides comprehensive services to support the development and application of in vitro protein evolution.
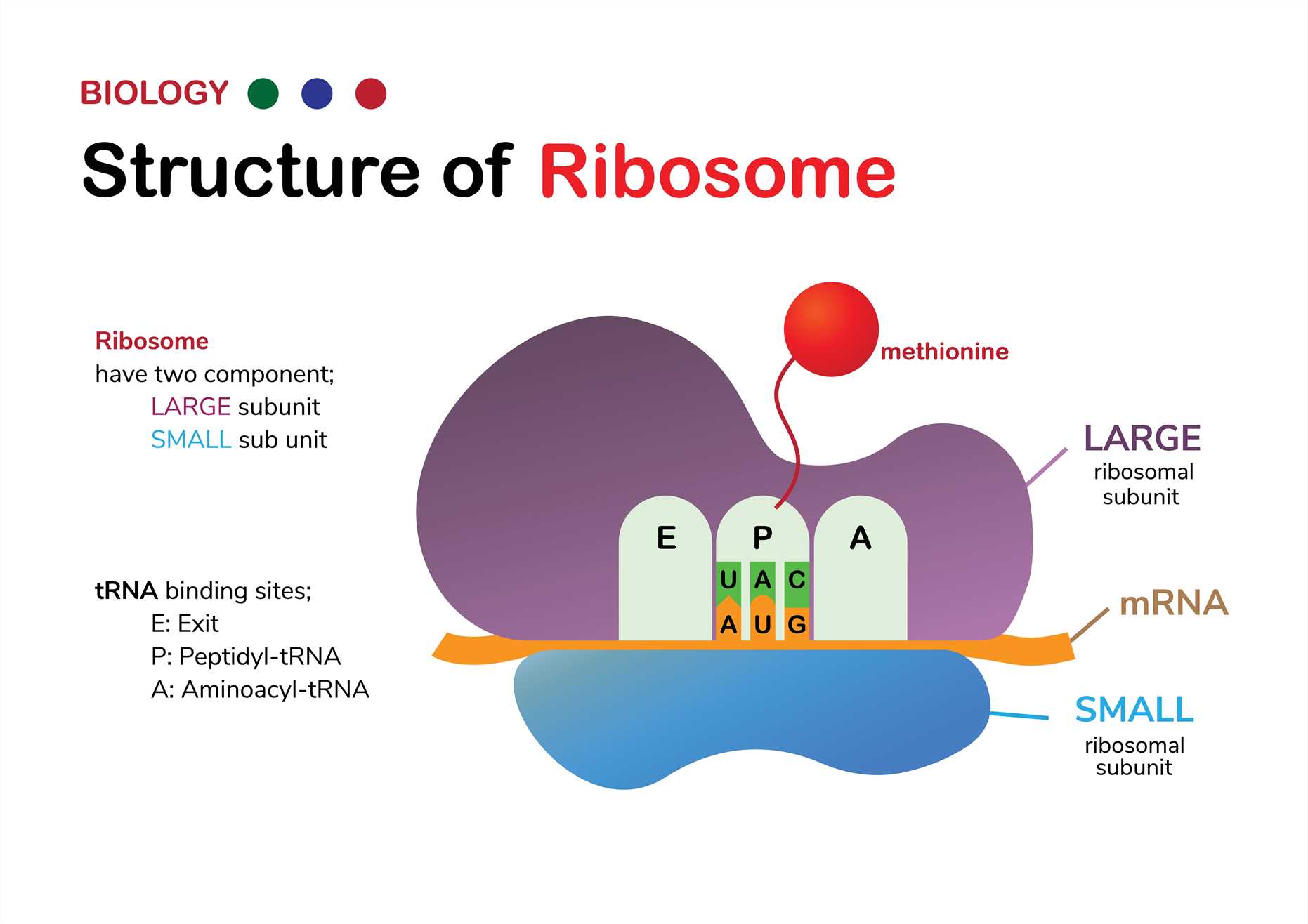 Fig 1. Structure of ribosome
Fig 1. Structure of ribosome
With the deepening of research, surface display technologies play an important role in the development and screening of proteins and antibodies, such as yeast display, phage display, bacterial display, and ribosome display systems. Compared with in vivo display technologies, ribosome display has some unique advantages. First, the size of the library is not limited by the transformation efficiency, which for E. coli is typically 109–1010 per microgram of DNA. When ligation mixtures are used, the transformation efficiency is significantly reduced. In ribosome display, the size of the library is limited only by the number of ribosomes in vitro translation, up to 1012–1014. Second, while protein biosynthesis and folding in living cells occur in a given environment, the use of an in vitro selection system allows the environment to be manipulated and optimized for the expression, folding, and stability of library members. This can be used for stability selection. Third, the diversity of library members can be easily manipulated during any selection step by introducing additional mutations using DNA shuffling and/or error-prone PCR. This is probably the most practical factor. Therefore, ribosome display is particularly suited for directed evolution projects over many generations.
Ribosome display technology has been applied to protein evolution in order to make them more potent in terms of stability, specificity, or affinity. A report investigated the generation of a variant library of M-MuLV reverse transcriptase (Moloney mouse leukemia virus) from the Moloney virus of mouse leukemia to identify the most active protein mutants. In the study, the ribosome displays were partitioned by emulsion (a method of personalizing each element in a population by means of a water-oil emulsion), so that each compartment represented only one variant in the library. In each compartment, the reverse transcriptase activity of selected mutant proteins was tested by detecting their ability to synthesize ADNc from mRNA. In this way, thermal stability and functional variants of proteins were identified and isolated. Another example of proteins showing evolution through the ribosome is the selection of functional enzyme variants. Several mutants of TEM-1, a β-lactamase associated with bacterial antibiotic resistance, were screened for specific inhibitors of bacterial enzymes (BLIP, β-lactamase inhibitory proteins) by ribosome display. Only one mutation with maintained enzyme activity was selected, thus escaping BLIP inhibition, validating the method of evolution and selection of ribosome display enzymes.
Creative Biolabs has a wealth of knowledge and experience in ribosome display. We would be happy to share with you our knowledge and experience in in vitro protein evolution.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.