Nuclear receptor superfamily comprises essential transcriptional regulators that play pleiotropic roles in cell proliferation, differentiation, development, and metabolism to govern digestive physiological and pathological processes. Many nuclear receptors (NRs) are ligand-activated transcription factors that regulate the expression of their target genes by affecting transcriptional activities and epigenetic alternations. With professional expertise in protein engineering areas, Creative Biolabs provides sophisticated computational pharmacology services for digestive and renal diseases based on structure-based NR modeling tools and protein interaction analysis.
Genetic and environmental factors that disrupt homeostatic digestive and renal activities could induce chronic kidney diseases or kidney failures, glomerulonephritis, Celiac & Crohn’s diseases, and others. Nuclear hormone receptors (NHRs), G-protein coupled receptors (GPCRs), and kinases expressed in digestive and renal tissues represent the most tractable targets for the development of competent drugs that can alleviate urological and metabolic disorders.
NHRs regulate lipid metabolism, carbohydrate metabolism, immune response, and inflammation. As a result of their diverse biological effects, they have become a major pharmaceutical opportunity for the treatment of metabolic diseases. Furthermore, several NRs (e.g. Vitamin D receptor (VDR), farnesoid X receptor (FXR)) activating ligands have shown a renal protective effect in the context of diabetic nephropathy.
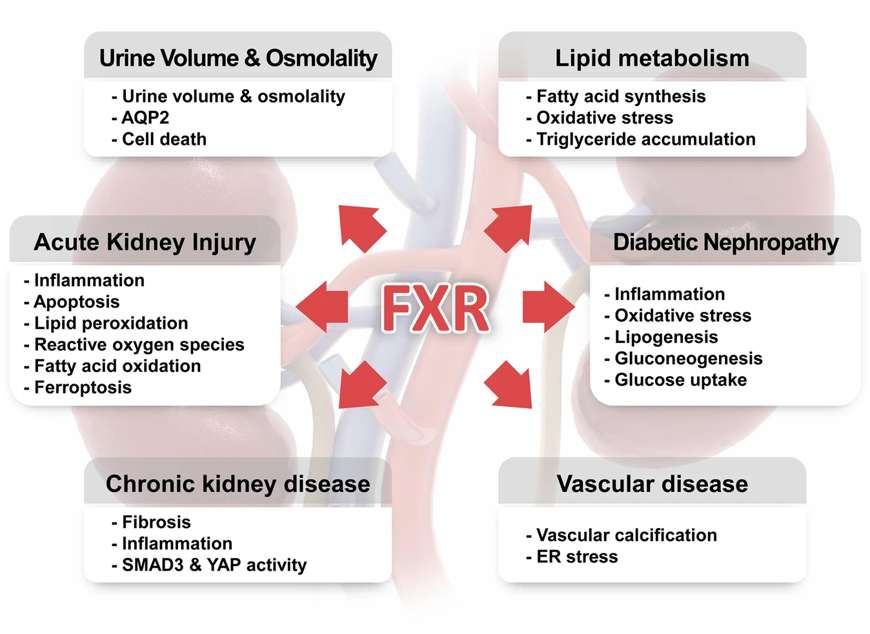 Fig.1 Roles of FXR in the physiology and pathophysiology of the kidney.1
Fig.1 Roles of FXR in the physiology and pathophysiology of the kidney.1
As ligand-dependent transcription factors, NRs can be activated by important molecules, such as endogenous hormones, glucocorticoids, steroidal hormones, and thyroid hormones. After activation, NRs will regulate the expression of specific genes and then participate in several pivotal processes, including development, homeostasis, and metabolism of the organism. Since NRs have effects on the expression of numerous genes associated with various digestive and renal abnormalities, they can be considered as an appropriate therapeutic target for novel drug discovery.
Renal PPARα takes a key part in modulating energy utilization in the kidney via the regulation of renal fatty-acid β-oxidation. The role of PPARγ agonists in preventing the development and progression of diabetic nephropathy has been reported. Notably, these two members of the PPAR subfamily have already been addressed by approved drugs.
Another example of a promising drug target for NRs is FXR which binds bile acids, the final product of cholesterol metabolism. Owing to the toxic properties of bile acids, their levels need to be tightly controlled. FXR signaling is involved in the regulation of intestinal bacterial flora, liver regeneration and, in case of misregulation, hepatocarcinogenesis.
Upon this situation, finding new bioactive scaffolds is a critical process in the field of drug design. At Creative Biolabs, we have not only identified NR target genes via experimental studies, but also developed in silico approaches to accelerate NR drug discovery for digestive and renal research. Our two-step modeling concept to identify active chemical compounds includes three-dimensional (3D)-pharmacophore filters and rescoring by shape alignment. Our computational analysis of 3D coordinates of protein structures has facilitated the detection of evolutionarily important residues and correlated them with important NR functions, like dimerization and DNA binding. As below, there is a summary of some relevant NRs that can be employed in structure-based modeling for digestive and renal diseases.
| ERRα | ERα | FXR | GR | PPARα |
| PPARγ | VDR | LXRα | LXRβ | PRα |
| PRβ | THα | THβ |
Now, potent computational approaches can be applied to detect NR target genes for drug development. As a first-class supplier in the protein design market, Creative Biolabs has developed a unique computer algorithm based on protein structure to predict NR target sequences in regulatory regions of genes. On the basis of different X-ray crystal structures, we’re able to help clients establish a virtual screening workflow for the identification of potential NR modulators for digestive and renal research. If you’re interested in our protein engineering services, please don’t hesitate to contact us.
Reference
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
