Creative Biolabs has been a leader in commercializing a comprehensive panel of methodologies in mapping both linear and conformational epitopes of monoclonal antibodies.
Epitopes consist of groups of amino acids that lie close together on the protein (antigen) surface and interact with an antibody; they therefore determine antigenicity. Epitopes of proteins are usually classified as either linear (continuous) or conformational (discontinuous). Conformational epitopes are thought to form the majority of epitopes on most proteins. However, many conformational epitopes may also be recognizable as linear epitopes.
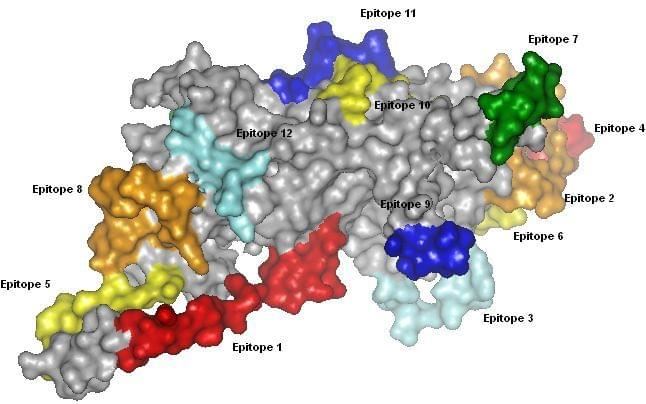
Figure 1: Predicted B cell epitopes of Human papillomavirus type 16 major late capsid protein L1
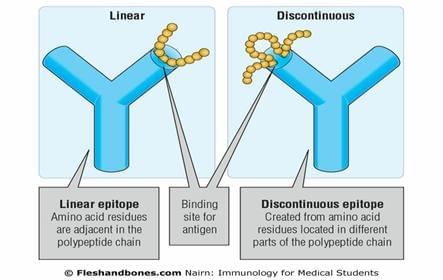
Figure 2: Linear and Conformational (discontinuous) epitopes
A multitude of disciplines require detailed knowledge about epitopes. Not only immunologists who have an a priori interest depend on epitope mapping protocols, but also biologists using antibodies as research tools, structural biologists studying protein–protein interactions, clinicians investigating patients' immune responses, vaccine developers designing and testing immunogens, diagnostic labs developing and applying ELISAs, and last but not the least, biotech and pharmaceutical companies are obliged to monitor the immunogenicity of novel therapeutic antibodies, proteins, and peptides, to mention only a few.
As epitope mapping is becoming more and more vital, technologies are developing fast too. Creative Biolabs has the unprecedented ability to provide epitope mapping services with 100% accuracy and efficiency. Creative Biolabs offers various types of epitope mapping technologies, among which you can choose based on the specific needs of your study.
Below are the methodologies that you can choose based on your specific needs:
Linear and conformational epitope mapping by crystallography
Staffed by dedicated scientists with combined expertise in antibody engineering and protein X-ray crystallography, Creative BioStructure has established an antibody epitope mapping platform based on protein crystallography. We have extensive experience in crystallizing antibody/antigen complexes and resolving their 3D structures. In particular, we are able to map the epitopes of a large variety of antibody forms, including intact IgG, Fab, scFv, F(ab')2, diabody, minibody, miniantibody, tandem scFv and single domain antibody as well as protein A scaffold. X-ray crystallography of antigen/antibody complexes allows mapping both conformational and linear epitopes.
Please refer to our website at Creative BioStructure for more information regarding resolving the structure of an antigen/antibody complex.
Linear epitope mapping by screening phage display peptide libraries
We rely on our phage display based Bio-fishing Platform to offer linear epitope mapping service. Of note, we have two random phage display peptide libraries that were constructed in house using Trimer Codon technology.
TriCo-16™ Phage Display Peptide Library
TriCo-20™ Phage Display Peptide Library
In addition, linear epitope mapping can also be performed using NEB Ph.D.TM-7 and Ph.D.TM-12 Phage Display Peptide Libraries.
NEB Ph.D.™-7 Phage Display Peptide Library
Ph.D.™-12 Phage Display Peptide Library
Compared with the commonly employed linear epitope mapping strategy of overlapping peptide arrays (chemically synthesized combinatorial peptide libraries), our phage display peptide libraries have a much greater diversity of the peptides and provide a method in which signals from individual peptides can be amplified for multiple rounds, thus significantly enhance the possibility of isolating the peptide of the perfect match.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.