Based on the optimized and validated surface display platform, Creative Biolabs provides clients high-class CreMap™ epitope mapping services to help avoid the pitfalls of variability in assays and benefit from the efficiency and accuracy of a dedicated facility.
Detailed information on epitopes is of great importance both for the understanding of immunological events and for the development of effective vaccine and diagnostic tools against a vast number of diseases. Among all the methods available, the surface display technology provides a powerful solution for epitope mapping due to their relative cheapness and quickness.
The key principle of display techniques is testing the binding ability of a variety of peptides presented on the display platforms to the antibody of interest. The antigen DNA is fragmented by enzymes, expressed on the surface of the display platform to generate a library. After that, the binding ability of the peptide displayed on the phage surface to the antibody would be tested and the epitope mapping would be performed. One of the exclusive features of surface display technique is that the peptide which is expressed by the fragment containing the epitope would be recognized by the antibody. Thus, the information of the DNA fragment will be obtained simultaneously by sequencing back to the DNA fragment.
Phage display is one of the most frequent and popular display methods. Construction of phage display peptide libraries represents a popular way for screening epitopes. This powerful approach involves fusion of the foreign DNA fragments with the filamentous phage gene coding coat protein (e.g. pIII, pVI, pVII, pVIII, and pIX). In the case of M13 filamentous phage display, the peptides of interest are more often ligated into pIII gene. There are around five copies of pIII gene in M13 filamentous phage. This format works well and the general selection may not fall short when we are seeking high-affinity binders. The incorporation of peptides does not inhibit the phage cycle.
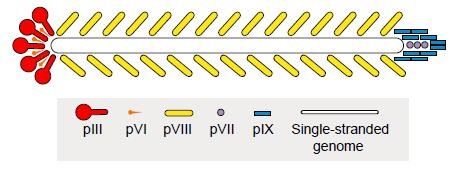 Fig.1 Schematic of the filamentous phage for recombinant display of foreign peptide.
Fig.1 Schematic of the filamentous phage for recombinant display of foreign peptide.
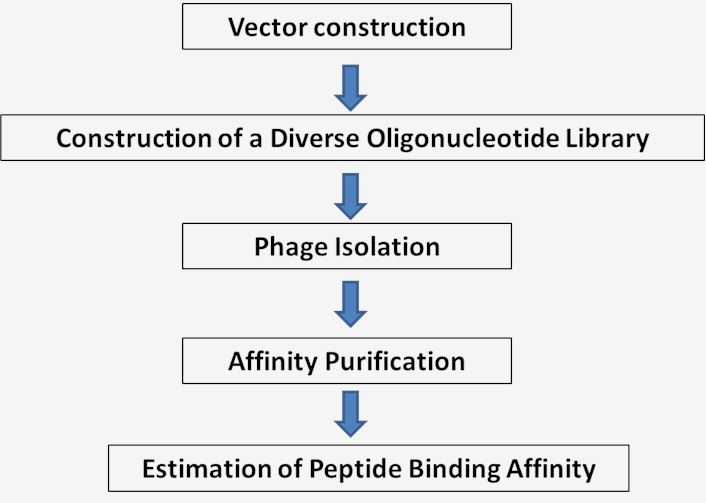 Fig.2 Workflow for surface display technique.
Fig.2 Workflow for surface display technique.
With extensive experience in epitope mapping service, Creative Biolabs is dedicated to helping clients determining epitopes with well-established surface display platform. If you are now doing research on antigen-antibody complex, trying to figure out epitopes on the antigen, or if you are interested in our surface display platform, please contact us and a formal feedback will be sent back as soon as possible.
Other optional CreMap™ B cell epitope mapping services:
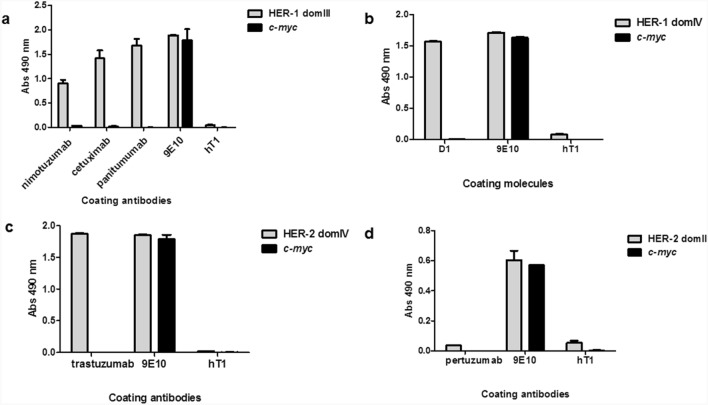 Fig. 3 Recognition of phage-displayed domains of HER-1 and HER-2 by monoclonal antibodies in ELISA. (Dayana Pérez-Martínez, 2022)
Fig. 3 Recognition of phage-displayed domains of HER-1 and HER-2 by monoclonal antibodies in ELISA. (Dayana Pérez-Martínez, 2022)
The research explored the potential of phage display technology for domain-level epitope mapping of polyclonal antibodies targeting HER-1 and HER-2 receptors, which are significant in the context of cancer. The study highlighted how polyclonal antibodies generated through immunization recognized multiple domains of HER-1 and HER-2 displayed on phages, reflecting the diversity of antibody responses. Importantly, the use of phage display facilitated the identification of how these antibodies recognize mutated receptor variants that commonly arise in cancer patients under selective therapeutic pressure. This method allows for detailed mapping of epitopes on a domain level, which is crucial for designing vaccines and therapies that can overcome resistance by targeting multiple epitopes or mutated antigens effectively.
Epitope mapping by surface display technique involves presenting peptide libraries or protein fragments on the surface of host cells or phage particles to identify regions recognized by antibodies. This method enables the comprehensive identification of epitopes through high-throughput screening of interactions between the displayed peptides and specific antibodies. It is a valuable tool for studying immune responses and designing targeted therapies.
The surface display technique works by expressing peptide libraries or protein fragments on the surface of host cells, such as bacteria, yeast, or phages. These displayed peptides interact with antibodies in a screening process. Positive interactions are detected using various methods, such as fluorescence or phage enrichment, allowing the identification of epitopes recognized by the antibodies. The epitopes are then sequenced and analyzed to determine their specific regions on the antigen.
The surface display technique offers several advantages for epitope mapping, including high-throughput capability, the ability to present large and diverse libraries of peptides, and the potential for identifying conformational epitopes. This method is cost-effective and allows for the direct selection of peptides that bind to antibodies with high affinity. Additionally, it can be applied to various host systems, providing flexibility in experimental design.
Epitope mapping by surface display technique benefits various applications, including vaccine development, therapeutic antibody design, and diagnostic tool creation. It aids in understanding immune responses, identifying immunodominant regions of pathogens, and discovering new therapeutic targets. This technique is also valuable in studying autoimmune diseases and developing personalized medicine approaches by identifying specific epitopes relevant to individual patients.
Epitope mapping by surface display contributes to identifying the precise regions of antigens that antibodies recognize and bind to. This information helps in designing antibodies that target specific epitopes, enhancing their efficacy and specificity. The technique also allows for the identification of potential cross-reactive epitopes, reducing the risk of off-target effects and improving the safety profile of therapeutic antibodies.
Traditional epitope mapping methods, such as peptide synthesis followed by ELISA or spot synthesis, are often labor-intensive, time-consuming, and less capable of identifying conformational epitopes. The surface display technique overcomes these limitations by enabling the presentation of a wide array of peptide libraries in their native conformations, allowing for the identification of both linear and conformational epitopes. It also provides a high-throughput and cost-effective solution.
Data from surface display epitope mapping is analyzed by sequencing the peptides or protein fragments that interact with the antibodies. After identifying the positive interactions, the sequences of these peptides are determined using techniques such as next-generation sequencing or Sanger sequencing. Bioinformatics tools are then used to align the sequences to the antigen, pinpointing the specific epitopes recognized by the antibodies and analyzing their characteristics.
Surface display technique can be adapted for epitope mapping of virtually any antigen. The peptides or protein fragments representing potential epitopes can be derived from the known or predicted sequence of the antigen. This versatility makes the technique applicable to a wide range of research areas, including infectious diseases, cancer, and autoimmune disorders, facilitating the detailed study of antibody-epitope interactions across different antigens.
The surface display technique improves the accuracy of epitope mapping by presenting peptides or protein fragments in their native conformations on the surface of host cells or phages. This allows for the identification of both linear and conformational epitopes. The high-throughput screening capability of this method ensures comprehensive coverage of the antigen, leading to precise and reliable identification of epitopes.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
