Creative Biolabs provides epitope mapping services by using limited proteolysis coupled with the most advanced mass spectrometry equipment. The enzymes used in our platform provide highly optimized digestion conditions with the highest sequence coverage possible. Creative Biolabs is confident to find the epitope region with high accuracy and resolution.
Accurately characterizing epitopes on an antigen is a critical step for understanding the pathogenesis of an infectious material, and for the development of drugs and vaccines against toxins or enzymes. Proteolytic fragmentation has been used for mapping protein epitopes of antigens and proteolytic cleavage of antigen-antibody complexes provides a simple, cost-effective and straightforward approach to characterize a linear epitope. Proteins are cleaved by enzymes with different restriction sites to generate overlapping peptides. These peptides are then separated and identified by Mass Spectrometry (MS). The application of MS to characterize peptides and proteins has become a versatile and powerful method to precisely determine a soluble antigen’s partial structure (epitope) which interacts with the antibody. By comparing free antigen and antigen-antibody complex, a linear epitope on the antigen can be accurately mapped in a short time. Over the years this method has been established as a reliable source of data for both research and therapeutic purposes.
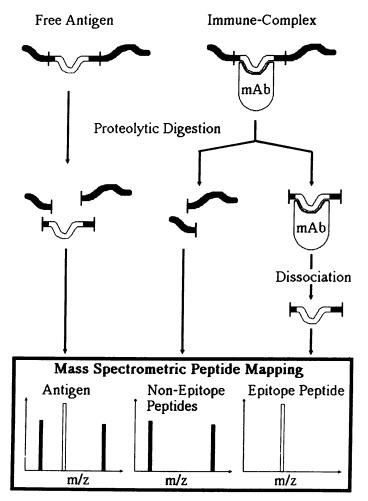 Fig.1 Scheme of the epitope mapping method by limited proteolysis.
Fig.1 Scheme of the epitope mapping method by limited proteolysis.
Creative Biolabs has accumulated extensive experience to offer epitope mapping services. Our experienced scientists will assist you in designing the research program that best suits your purpose and advances your research. If you are now trying to figure out epitopes on an antigen, or if you are interested in our platform, please contact us. A formal feedback will be sent back as soon as possible. We are always more than ready to reach out.
Other optional CreMap™ B cell epitope mapping services:
Epitope mapping by limited proteolysis and mass spectrometry involves digesting proteins with specific proteases under controlled conditions, then identifying the resulting fragments using mass spectrometry. This method helps pinpoint protein regions (epitopes) that are involved in interactions with antibodies or other molecules, based on which fragments are protected or exposed.
Limited proteolysis selectively breaks down protein regions that are accessible and flexible, leaving structured or bound regions (like those involved in binding to antibodies) intact. Analyzing which parts of the protein are digested and which are not provides clues about where epitopes are located on the protein surface.
Mass spectrometry accurately identifies and quantifies the peptides resulting from proteolysis. It measures the mass-to-charge ratio of ions to identify the molecular structure of peptides, helping researchers determine which parts of the original protein were protected due to binding interactions.
This technique is highly sensitive and can provide detailed information about the primary structure of protein epitopes. It is effective even with complex mixtures of proteins and can distinguish between closely related peptides, offering high-resolution insights into protein-protein interactions.
Limited proteolysis coupled with mass spectrometry can differentiate between linear and discontinuous epitopes. By examining the pattern of peptide fragments, researchers can infer whether epitopes are composed of contiguous amino acid sequences or if they are made up of segments brought together by protein folding.
This method may not effectively identify epitopes that involve large or complex conformational structures unless those structures influence the protein's susceptibility to protease digestion. Additionally, the interpretation of mass spectrometry data requires sophisticated software and expertise in protein chemistry.
Compared to methods like X-ray crystallography or NMR spectroscopy, limited proteolysis and mass spectrometry is less labor-intensive and does not require crystallization or isotopic labeling. However, it provides less direct information about the spatial arrangement of atoms within the epitope compared to these more direct structural methods.
Recent improvements include the use of more specific and varied proteases to achieve finer control over digestion, as well as advances in mass spectrometry technology that improve sensitivity and accuracy. Enhanced computational tools for analyzing mass spectrometry data have also been developed, helping to better predict and characterize epitopes.
Beyond epitope mapping, limited proteolysis and mass spectrometry are used in studying protein folding, protein-protein interactions, and protein stability. It's also utilized in quality control for therapeutic proteins and vaccines to ensure that correct structural conformations are maintained, which is crucial for their efficacy and safety.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
