Glycosylated Exosomal Proteomic Detection Service
Overview Services Features FAQs
Creative Biolabs offers a high-throughput detection service of glycosylation-modified proteins with low contents and a wide dynamic range in exosomes through mass spectrometry and enrichment processing.
Introduction of Glycosylated Proteins
-
In eukaryotes, more than half of the proteins can be glycosylated to become glycoproteins, 90% of which are N-glycoproteins. Glycosylation involves a wide range of functions: (i) protection, stabilization, and barrier to cells, (ii) specific ligands for exogenous receptors, and some glycoproteins can act as specific receptors for various viruses, bacteria, and parasites, (iii) glycoproteins can also act as specific ligands for endogenous receptors, mediating clearance, turnover, and intracellular penetration, (iv) glycoproteins also play an important role in the fertilization process.
-
Glycosylation modifications are divided into two broad categories: N- and O-glycosylation
-
N- glycosylation: refers to glycoproteins formed by covalent linkage of the sugar chain to the amino group on the protein aspartate, which tends to have the amino acid sequence Asn-X-Thr/Ser/Cys (X≠Pro) with a classical core pentasaccharide structure (Man3GlcNAc2).
-
O- glycosylation: Linked to the hydroxyl groups of Ser, Thr, hydroxylysine (collagen), or Tyr (glycogenic protein) via glycosidic bonds, which typically are shorter and without a core structure. Given this, the resolution of the O-glycosylated glycan structure is more difficult than that of the N-glycosylation.
-
Meanwhile, glycosylation modification has macroscopic and microscopic heterogeneity, as the same glycan can modify different sites of the same protein, and the same site can also modify other structures of the glycan, which is very difficult to resolve and becomes a complex problem in glycosylation research.
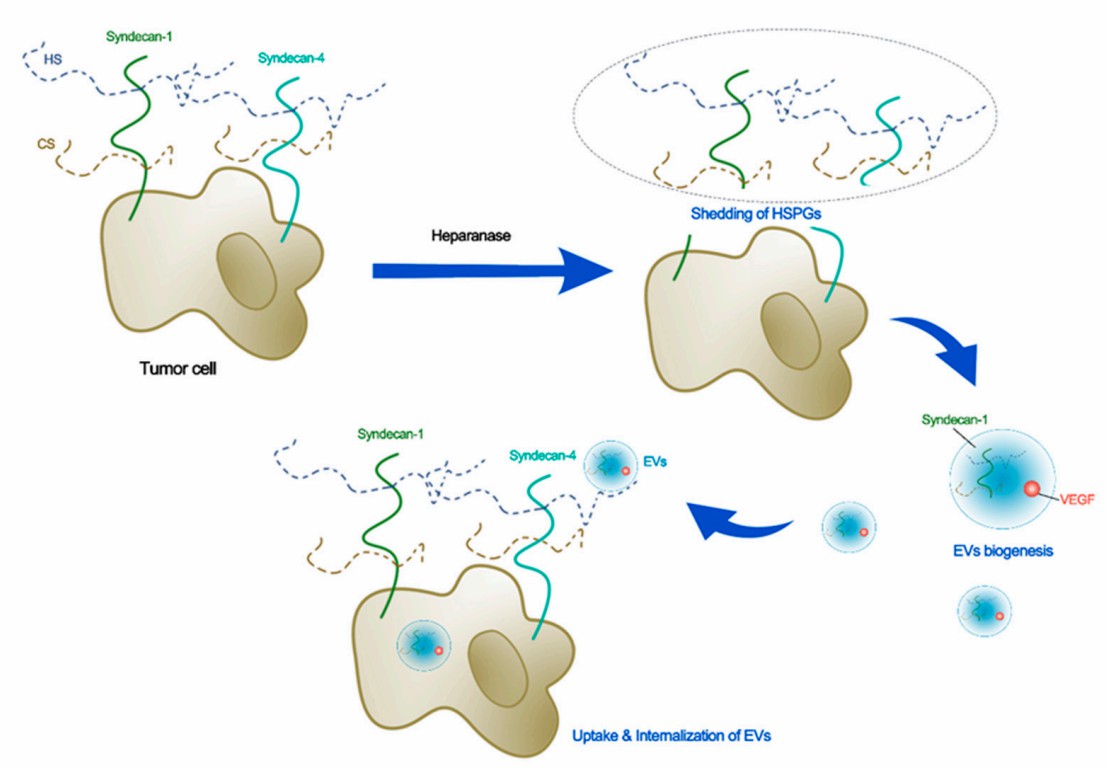 Fig.1 Schematic for the potential roles of glycocalyx in biogenesis, uptake, and internalization of EVs.1
Fig.1 Schematic for the potential roles of glycocalyx in biogenesis, uptake, and internalization of EVs.1
Glycosylated Exosomal Proteomic Detection Service at Creative Biolabs
Traditional platforms for glycoprotein discovery generally excise glycans from peptides by glycosidases (either retaining the peptide or the glycan chain), and then obtain glycosylation modification sites on the peptides by mass spectrometry analysis or resolve the glycan chain structure. These strategies lose the information on glycan structure and peptide sites, which fail to achieve the correspondence analysis between glycan structure and peptide sites, missing the core information of glycosylation proteomics study.
At Creative Biolabs, exosomal proteins are cleaved into peptides using enzyme specificity and glycopeptides are obtained by enrichment methods. The glycopeptides are then fragmented by stage using high-energy collision fragmentation mass spectrometry, to obtain ion information at all levels of the mass spectrum. The comparative analysis of glycosylation sites and glycoconjugate structures of exosomal peptides is performed by mass spectrometry data comparison software and a self-built glycopeptide database, which can retain glycosylation site information as well as resolve the structure of glycans, providing unique advantages in comparative glycosylation histology studies. Our glycosylated exosomal proteomics service will deliver detailed reports including
-
Glycosylated exosomal protein processing and profiling procedures.
-
Raw data collected by the mass spectrometer.
-
Comprehensive identification data of complete N-glycopeptide IDs based on target-decoy library search and spectrogram level FDR control, including peptide backbone (amino acid sequence, potential N-glycosylation sites) and N-linked glycan fraction (monosaccharide composition, potential sequence, and glycosidic bond linkage structure). Among them, the N-glycosylation sites are further confirmed by site-determining fragmentation ions, and the sequence and linkage structures are further confirmed by structural diagnostic fragmentation ions.
-
Results of differential proteins bioinformatics analysis.
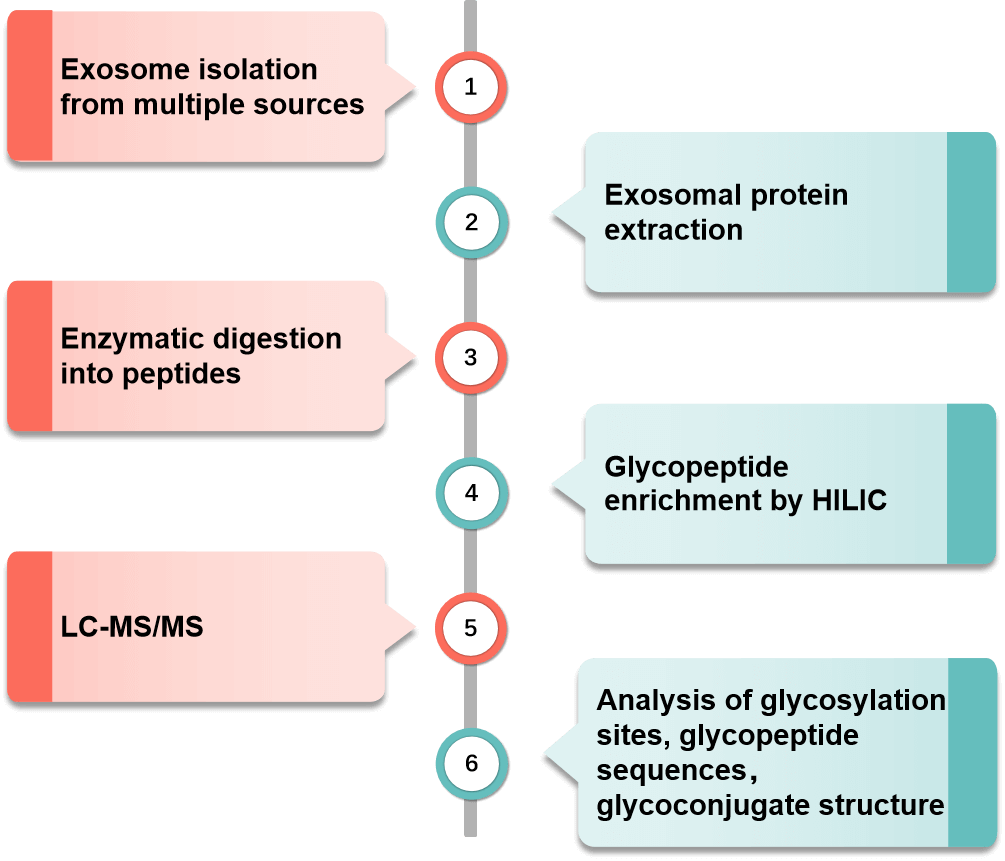 Fig.2 Glycosylated exosomal proteomic detection workflow.
Fig.2 Glycosylated exosomal proteomic detection workflow.
Features of Glycosylated Exosomal Proteomic Detection Service
-
Advanced Technology: Our service utilizes high-resolution nano LC-MS/MS technology for acquiring precise glycosylated protein profiles of exosomes.
-
Comprehensive Analysis: We provide a comprehensive analysis of glycosylation patterns, including N-linked, O-linked, and other complex carbohydrates attached to proteins.
-
Accurate Detection: Our detection service specializes in using lectin affinity chromatography and other precise methods for accurate glycoprotein identification and quantification.
-
High Sensitivity: We employ highly sensitive techniques for detecting low abundance and subtle glycosylation changes that can be informative for disease states.
FAQs
Q: How sensitive is your glycosylated exosomal proteomic detection service?
A: Our service utilizes advanced technology, providing high sensitivity and specificity. Even low-abundance proteins and subtle alterations in glycosylation can be accurately detected.
Q: Can this service detect different types of glycosylation?
A: Yes, our service can accurately analyze different types of glycosylation, including N-linked, O-linked, and complex carbohydrate modifications.
Q: What kind of equipment do you use for detection?
A: We employ advanced analytical methods like nano LC-MS/MS technology, and lectin affinity chromatography for precise detection and quantification.
Creative Biolabs offers a specialty glycosylated exosomal proteomic detection service to advance research on the biological significance associated with exosomal protein glycosylation modifications. Please contact us with your interest.
Reference
-
Zeng, Ye, et al. "Glycocalyx acts as a central player in the development of tumor microenvironment by extracellular vesicles for angiogenesis and metastasis." Cancers 14.21 (2022): 5415. Under Open Access license CC BY 4.0. The image was modified by revising the title.
For Research Use Only. Cannot be used by patients.
Related Services:

 Fig.1 Schematic for the potential roles of glycocalyx in biogenesis, uptake, and internalization of EVs.1
Fig.1 Schematic for the potential roles of glycocalyx in biogenesis, uptake, and internalization of EVs.1
 Fig.2 Glycosylated exosomal proteomic detection workflow.
Fig.2 Glycosylated exosomal proteomic detection workflow.









