Creative Biolabs has established a series of technical platforms that monitor and detect protein-protein, protein-DNA, and protein-RNA interactions, particularly yeast hybrid systems. Among them, yeast one-hybrid (Y1H) assay is increasingly attracting more attention in analyzing the interaction between protein and nucleic acid. The Y1H is usually used to identify protein-DNA interactions (PDIs) with short cis-regulatory elements and with longer and more complex DNA fragments, for instance, promoters and enhancers. Creative Biolabs provides Y1H services including Y1H library construction and screening.
Precise gene expression plays a pivotal role in cell development, homeostasis, as well as in response to environmental cues. It is largely controlled by the specific binding of heterologous transcription factors (TFs) to the regulatory DNA sequence. TFs consist of 5-10% of the protein-coding genes in most organisms, also referred to the sequence-specific DNA-binding factor. They are actually proteins responsible for regulating transcriptional rates of genetic information from DNA to mRNA. PDIs can be experimentally identified by many different approaches, and the Y1H assay is regarded as a gene-centered method to confirm the repertoire of TFs that bind a DNA region of interest.
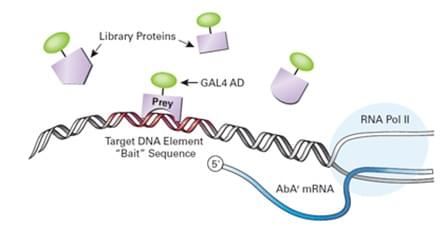 Fig.1 Design and application of Y1H system.1
Fig.1 Design and application of Y1H system.1
Y1H is a powerful approach to rapidly identify proteins that can interact with a specific DNA regulatory region of interest. This technique is employed to ascertain “prey” molecules of TFs bind to a DNA fragment of interest that is known as “bait”. The process of Y1H involves that plasmids encoding TFs introduced into a yeast bait strain in which the DNA fragment is integrated upstream of one or more reporter genes, and subsequently the activation of the reporters indicates that a TF-DNA interaction has occurred. The name of one hybrid in yeast is derived from these plasmids, which express each TF as a hybrid protein fused to the activation domain (AD) of yeast Gal4.
In this assay, there are two major components included. One, a reporter is constructed in which a bait DNA is cloned upstream of reporter genes, and the other, a plasmid that expresses a prey hybrid protein comprises a TF fused to the AD of the yeast TF Gal4. Both constituents are introduced into a budding yeast strain, and the bait part is used to “fish” for the interacting prey. If the hybrid TF binds the DNA of interest, the AD will activate the expression of reporter genes. In conclusion, Y1H is an accessible technology to detect PDIs between TFs fused to the Gal4 AD (AD-TF “prey”) and a specific DNA sequence (DNA “bait”).
 Fig.2 Princple of modified Y1H system for transcription factors selection.2
Fig.2 Princple of modified Y1H system for transcription factors selection.2
Y1H is based on the interaction of a TF with a DNA fragment upstream of reporter genes. Multiple reporters have been served to detect the interaction in Y1H assays, such as auxotrophic genes of HIS3, LEU2, URA3, and TRP1 that enable growth in the absence of histidine, leucine, uracil, and tryptophan, respectively, as well as LacZ that codes for the bacterial enzyme beta-galactosidase, which is detectable in colorimetric tests.
Although Y1H results are subject to false-negative or false-positive interactions as with any methodology, there are several advantages cannot be negligible as follows.
Ultimately, the comprehensive detection of PDIs that drive gene regulation requires a combination of complementary means, such as chromatin immunoprecipitation (ChIP). Remarkably, our combination of the one-hybrid and two-hybrid approaches can increase the stringency and reliability of results obtained from two-hybrid assays alone. With the assistance of Creative Biolabs, scientists around the world will dissect more information about protein-nucleic acid interactions and facilitate their understanding of gene expressions and regulations.
Other optional protein-nucleic acid interaction (PNI) assay services:
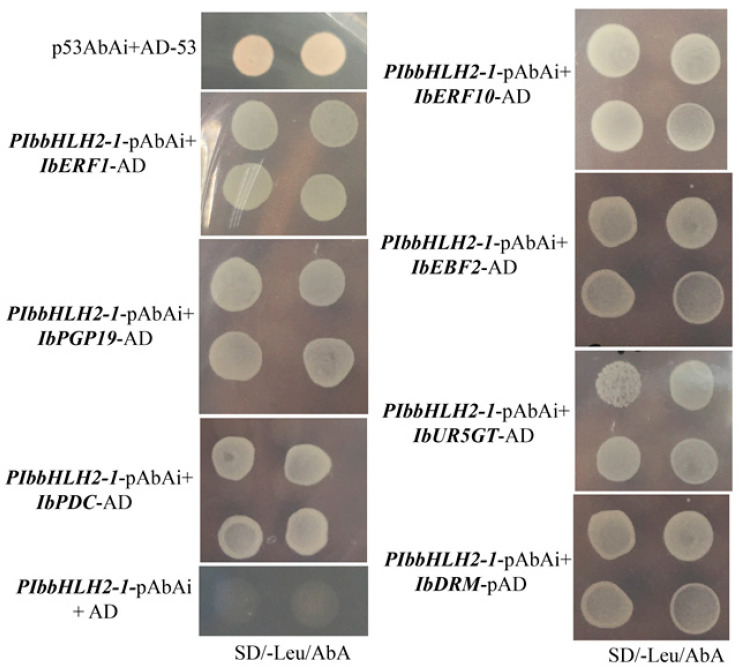 Fig.3 Interaction between transcription regulators and pibbhlh2-1 determination via Y1H.3
Fig.3 Interaction between transcription regulators and pibbhlh2-1 determination via Y1H.3
Some studies have shown that transcription factor IbbHLH2 is involved in the anthocyanin biosynthesis of purple sweet potatoes, but little is known about the role of transcriptional regulators upstream of the IbbHLH2 promoter in anthocyanin biosynthesis. Here, seven proteins—IbERF1, IbERF10, IbEBF2, IbPDC, IbPGP19, IbUR5GT, and IbDRM—were selected from purple sweet potato tubers by the yeast one-hybrid assay as upstream binding proteins of the IbbHLH2 promoter. At the same time, they used a double luciferase reporter gene and the yeast two-hybrid assay to verify the interaction between the promoter and these upstream binding proteins. In addition, they analyzed the expression levels of transcriptional regulatory factors, transcriptional factors, and structural genes related to anthocyanin synthesis in purple and white sweet potato roots at different root development stages by real-time fluorescence quantitative PCR. The results showed that IbERF1 and IbERF10 were key transcriptional regulators of the IbbHLH2 promoter.
Drosophila pigmentation is a model system for studying the evolutionary changes in gene regulation. The differences in the expression of these genes are related to different pigment patterns among species, and this difference has been proven to be caused by changes in cis- and trans-regulation. Through yeast one-hybrid assay and RNAi screening, the researchers looked for transcription factors that bind to yellow cis-regulatory sequences and affect abdominal pigmentation in Drosophila melanogaster. Through yeast one-hybrid assay, they screened out 45 direct regulators that may have yellow expression. In the RNAi screens, 32 regulatory factors were shown to be associated with abdominal pigmentation. In the two screenings, a total of nine transcription factors were selected, including four nuclear receptors related to ecdysone signal transduction (Hr78, Hr38, Hr46, and Eip78C). This finding suggests that the yellow expression of nuclear receptors that may be directly affected by ecdysone is controlled during early pupa development (when adult pigments are formed).
It is especially suited for identifying and characterizing interactions between DNA-binding proteins and their corresponding DNA sequences. This makes it a valuable tool in studies aimed at mapping gene regulatory networks, understanding transcriptional regulation, and characterizing the function of transcription factors. It is also useful in identifying novel DNA-protein interactions when exploring the regulatory mechanisms of less characterized or newly discovered genes. The Y1H assay can be applied in functional genomics to associate specific proteins with particular DNA motifs to infer potential regulatory roles.
Interpreting results from a Yeast One-Hybrid (Y1H) assay involves analyzing the activation of the reporter gene in response to the interaction between the DNA-binding protein and the DNA sequence of interest. Positive results, indicated by the expression of the reporter gene (such as growth on selective media or color change in a colorimetric assay), suggest that the protein can bind to the DNA sequence. However, it's important to confirm these results through additional experiments:
Quantification in Y1H assays can be achieved by using reporter genes that produce measurable outputs, such as fluorescent or luminescent reporters, which allow for the quantification of interaction strength. This is particularly useful for comparing the relative affinities of different proteins for the same DNA sequence or analyzing the effects of mutations within the protein or the DNA on binding efficiency. Quantitative Y1H assays require careful calibration and normalization to control for variations in expression levels and yeast cell density, ensuring that the measurements reflect true differences in protein-DNA interaction strengths.
Different strains may have variations in their transcriptional machinery, affecting the basal levels of reporter gene expression and the overall background noise. Some strains are engineered to have lower auto-activation, making them more suitable for detecting weak interactions without significant background. Others might be modified to improve the overall health and growth rate under assay conditions, enhancing the reproducibility and reliability of results. Selecting an appropriate yeast strain is crucial for minimizing false positives and ensuring that the detected interactions are as biologically relevant as possible.
Yeast one-hybrid (Y1H) assays can be automated to some extent, especially for high-throughput screening purposes. Automation involves using robotic systems and liquid handling devices to prepare and dispense yeast cultures, transform yeast cells with DNA, and handle reporter gene assays. This automation is particularly useful in large-scale studies where numerous protein-DNA interactions need to be tested. Automated systems can significantly speed up the process, increase throughput, and reduce human error, making it feasible to screen extensive libraries of proteins and DNA sequences systematically.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
