Creative Biolabs is a leading service provider of peptide repertoire analysis. We have developed an advanced Magic™ analysis platform for the identification of peptide repertoire. Combined with our phage display platform, we are able to provide custom peptide binder selection and epitope mapping service tailored to the unique requirements by our customers.
Phage display technology has been widely used in peptides library selection for the identification of peptide ligands and epitope mapping. The screening consists of multiple rounds of affinity selection and the phage amplification and then followed by ELISA-based selection and sequencing of the positive clones. Furthermore, a serial of peptides will be synthesized and tested for their binding to the targets. A key step of the screening process is the analysis of the sequences and the determination of the consensus motifs. With the same consensus motif, peptides may bind to the same epitope on the target by similar molecular interactions. Different consensus sequences may suggest different binding modes on the same or different surface areas.
In drug development, multiple consensus sequences are highly preferred to develop different peptide leads for success. However, the conventional Sanger sequencing is limited due to its low throughput feature. The homology information of the peptides could hardly be discovered by the traditional sequencing method.
The Magic™ peptide repertoire analysis service is qualified to analyze the abundance and diversity of peptides selected from phage display library. We have developed an optimized sequencing analysis method which reduces the biases and the false positives arise from the traditional sequencing method. The Magic™ analysis could identify the rare peptide motifs with binding ability, and also precisely define the consensus sequences and the sub-groups. With the information, the peptide leads could be identified for drug development and also the epitopes could be identified in an efficient manner. Even a single round of phage selection may be enough for binding motif selection. Such an analysis method could avoid the propagation-related biases, reducing the procedures of bacterial infection, phage propagation and purification.
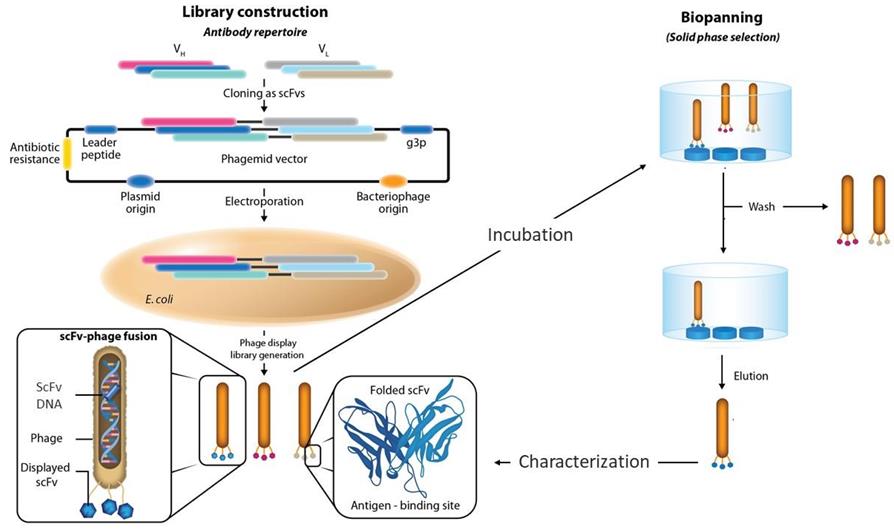 Fig.1 Phage display libraries for antibody development.1
Fig.1 Phage display libraries for antibody development.1
Creative Biolabs is a well-recognized expert at phage display peptide library services. During the past decade, our scientists have assisted many customers to accomplish their peptide binder identification or epitope mapping projects. With the Magic™ peptide repertoire analysis platform, our proprietary protocol and tailored analysis strategy guarantee the generation of peptides with desired features.
Other optional Magic™ therapeutic antibody discovery services:
Peptide repertoire sequencing analysis is used to study the diversity and composition of peptides, often derived from proteolytic cleavage of proteins, within a biological sample. This method involves high-throughput sequencing of peptide fragments to understand the range of peptides present, their origin, and their potential roles in biological processes. It is commonly used in immunology, oncology, and drug discovery to explore peptide-based interactions and immune responses.
Traditional proteomics typically involves identifying and quantifying proteins within a sample, whereas peptide repertoire sequencing focuses specifically on the diversity and sequence composition of peptides derived from proteins. This method provides a more detailed view of the specific peptide fragments generated through proteolysis, which can be critical in understanding immune recognition, antigen presentation, and peptide-based therapeutics. Peptide repertoire sequencing also allows for the exploration of post-translational modifications and variant peptides.
Peptide repertoire sequencing analysis is used in identifying neoantigens for cancer immunotherapy, studying antigenic peptides presented by MHC molecules in immunology, and exploring peptide-based drug discovery. It is also valuable in understanding autoimmune diseases by analyzing the peptides that trigger immune responses and in infectious disease research by identifying pathogen-derived peptides that elicit immune reactions.
Peptide repertoire sequencing can identify tumor-specific peptides, known as neoantigens, which arise from mutations in cancer cells. These neoantigens can be used to develop personalized cancer vaccines or as targets for T-cell-based therapies. By analyzing the peptide repertoire presented by MHC molecules on tumor cells, researchers can discover unique peptide signatures that differentiate cancer cells from normal cells, leading to more effective and targeted immunotherapies.
In immunology research, peptide repertoire sequencing is used to study the peptides presented by MHC molecules on the surface of cells, which are critical for immune recognition. By sequencing these peptides, researchers can identify the specific antigens that are recognized by T cells, understand immune responses to pathogens, and explore the mechanisms of autoimmune diseases. This information can guide the development of vaccines, immunotherapies, and diagnostics.
Peptide repertoire sequencing analysis can detect post-translational modifications (PTMs) such as phosphorylation, glycosylation, and ubiquitination. By analyzing the sequence data, researchers can identify peptides that have undergone PTMs and study their roles in biological processes. Detecting PTMs is important for understanding protein function, regulation, and the mechanisms underlying diseases, as well as for identifying novel therapeutic targets.
Peptide repertoire sequencing analysis has the ability to identify and characterize bioactive peptides with therapeutic potential. By analyzing peptide repertoires, researchers can discover naturally occurring peptides that modulate biological pathways, serve as signaling molecules, or interact with specific receptors. This method also enables the identification of peptide-based inhibitors or mimetics that can be developed into drugs, offering a pathway to novel therapeutics with high specificity and low toxicity.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
