Analysis of the human leukocyte antigen (HLA) type is an important component of immunological research and biopharmaceutical research. Traditionally, micronucleotoxicity (MLCT) and flow cytometry (FC) were widely used to detect HLA types. Various molecular biology methods are also employed because of the limitations and deficiencies of MLCT and FC, while this expensive option is somehow difficult for cash-strapped laboratories to afford. To help scientists solve this dilemma, Creative Biolabs has standardized the sequence-specific primer (PCR-SSP) technology and applied it to HLA type detection, which can be determined by electrophoretic analysis.
HLA generally refers to the entire leukocyte antigen system, which is controlled by genes on the short arm of chromosome 6, and is part of the genetic region known as the major histocompatibility complex (MHC). The physiological role of HLA is mainly to control the self-identification of the immune system and resist the invasion of microorganisms. HLA gene ensures the accurate judgment and attack ability of the body's external threats by virtue of its extreme diversity. Because some HLA antigens are recognized on all body tissues (rather than just blood cells), the identification of HLA antigens is described as “Tissue typing”. Traditionally, HLA antigens have been classified and defined by serological techniques that rely on obtaining live lymphocyte preparations and appropriate antisera to identify the availability of HLA antigens. In the past few years, DNA technology has been widely used and gradually replaced the serological technology in clinical applications in the field of histocompatibility and immunogenetics. Many laboratories have begun to develop and apply several different DNA methods for DNA typing to detect HLA alleles.
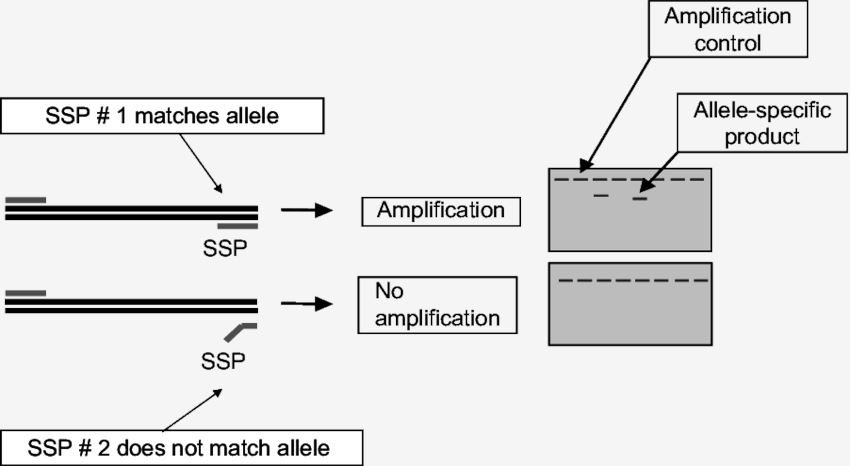 Fig.1 Principle of PCR-SSP.
Fig.1 Principle of PCR-SSP.
The PCR-SSP technique first appeared in the early 1990s and was based on the amplification of refractory mutation systems (ARMS). The principle of this method is that a perfectly matched primer is more efficient in a PCR reaction than one or more mismatched primers. Specificity is determined by the use of sequence-specific primers, where a 3' single-base mismatch inhibits the initiation of a non-specific reaction. Due to the lack of 3' to 5' exonuclease activity of Taq polymerase, even primer pairs do not specifically anneal and are not efficiently amplified. Therefore, only the required allele will be amplified, and then the amplified product can be detected by agarose gel electrophoresis.
Creative Biolabs has developed PCR-SSP technology to provide HLA typing services to a wide range of researchers. This method costs far less than traditional analytical methods such as flow cytometry, allowing you to get experimental results in the short term and make your project more efficient.
If you have any questions about our vector design service, you can contact us by email or send us an inquiry to find a complete solution.
Other optional ECIA™ HLA tissue typing service:
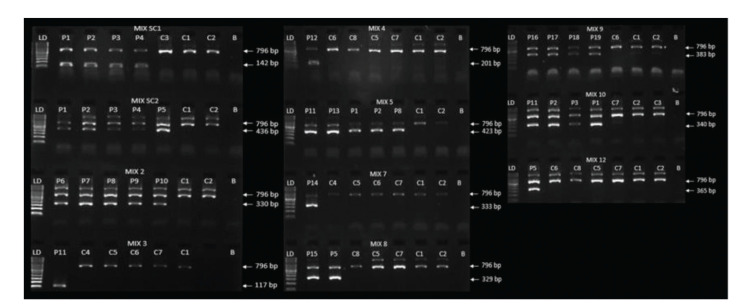 Fig. 2 Band pattern of positive and negative samples for HLA-B*27 allelic variants for all standardized PCR-SSP. (Fernanda Formaggi Lara-Armi, 2020)
Fig. 2 Band pattern of positive and negative samples for HLA-B*27 allelic variants for all standardized PCR-SSP. (Fernanda Formaggi Lara-Armi, 2020)
The article details the optimization of the HLA-B27 allele genotyping using the PCR-SSP (Sequence-specific Primer) method, emphasizing its application in diagnosing and managing ankylosing spondylitis (AS). The study validates the PCR-SSP technique against the more complex PCR-SSOP method, confirming high concordance rates and suitability for clinical diagnostics. This PCR-SSP method proves effective and reliable for heterozygous samples but has limitations in detecting certain homozygous conditions. The application of HLA Tissue Typing using the PCR-SSP method in this study involves specific, rapid genotyping of the HLA-B27 gene, crucial for confirming diagnoses of AS, thereby enabling timely and appropriate therapeutic interventions. This method provides a cost-effective, accessible option for routine clinical use, ensuring broader availability of essential diagnostic testing for AS.
PCR-SSP (Polymerase Chain Reaction with Sequence-Specific Primers) based HLA tissue typing is a method used to determine the human leukocyte antigen (HLA) alleles of an individual by amplifying specific HLA gene sequences. This technique is highly specific because it uses primers that only bind to particular sequences within the HLA genes of interest.
PCR-SSP enhances the accuracy of HLA tissue typing by using multiple sets of primers that are specific to distinct HLA alleles, allowing for precise amplification and identification of these alleles. This specificity reduces the risk of cross-reactivity and false-positive results, making it a reliable method for identifying HLA mismatches in transplant compatibility testing.
The main advantages of using PCR-SSP for HLA typing include its rapid turnaround time, cost-effectiveness, and the minimal amount of DNA required. Additionally, PCR-SSP can be performed in a straightforward manner without the need for sophisticated equipment, making it accessible for laboratories with limited resources.
PCR-SSP is versatile and can be used for typing most HLA class I and II alleles. However, its effectiveness may vary depending on the specific alleles and the primer sets available. It is particularly useful for high-resolution typing where specific allele identification is crucial.
The procedure involves extracting DNA from a blood or tissue sample, followed by PCR amplification using sequence-specific primers for the HLA alleles of interest. The resulting PCR products are then analyzed through gel electrophoresis to determine the presence or absence of specific alleles based on the size of the amplified fragments.
PCR-SSP is generally faster and less costly than PCR-SBT (Sequence-Based Typing), but PCR-SBT offers higher resolution and can identify new or rare alleles that PCR-SSP might miss. PCR-SSP is best suited for applications requiring rapid results and known allele screening, while PCR-SBT is preferable for comprehensive allele mapping and discovery.
Improvements in PCR-SSP techniques include the development of more specific and robust primer sets, automation of the PCR setup and analysis processes, and integration with digital platforms for data interpretation. These advancements have improved the throughput, accuracy, and user-friendliness of PCR-SSP based HLA typing.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
