Exosomal ceRNA Microarray Service
Overview Services Features FAQs
Long ncRNAs, circular RNAs, pseudogenes, miRNAs, and messenger RNAs (mRNAs) form a competitive endogenous RNA (ceRNA) network that plays an essential role in cancer, cardiovascular, neurodegenerative, and autoimmune diseases. After years of concentrating on exosomal RNA, Creative Biolabs introduces effective ceRNA microarray services to support our clients' exosomal ceRNA research.
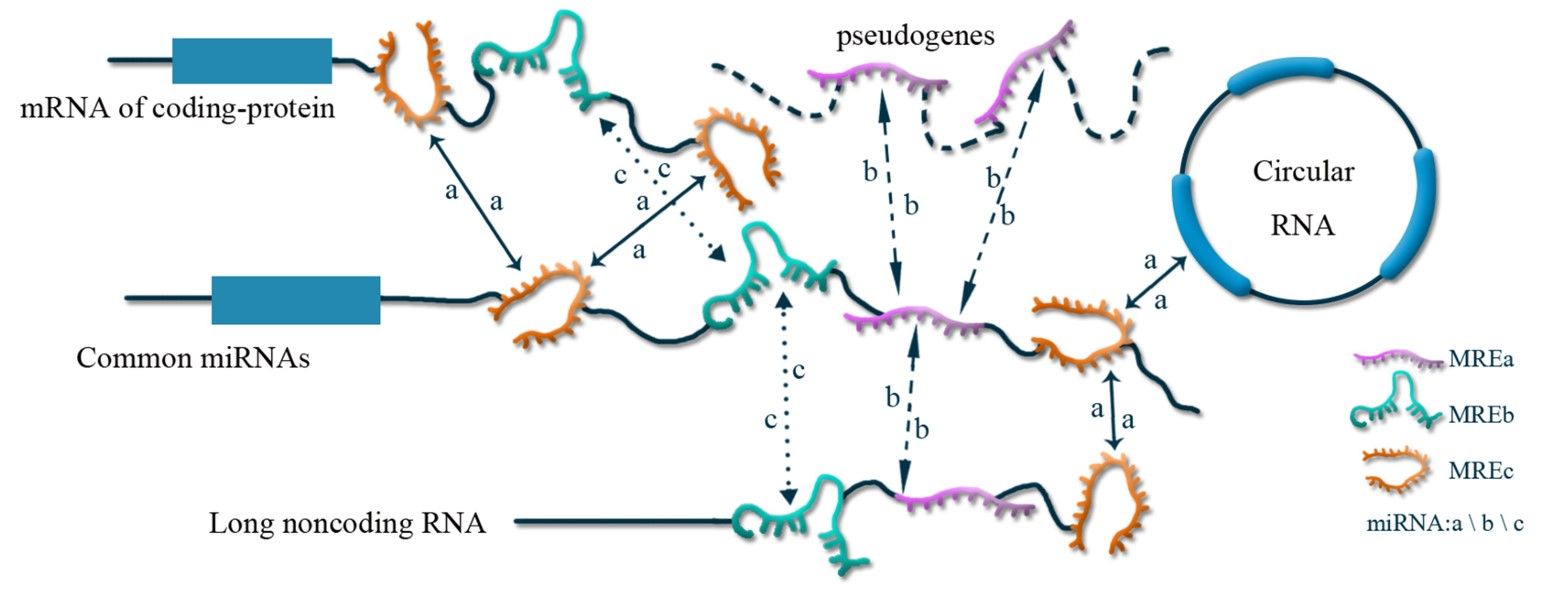 Fig. 1 The regulatory mechanism of exosomal ceRNA.1
Fig. 1 The regulatory mechanism of exosomal ceRNA.1
ceRNA in Exosomes
Emerging evidence has revealed that exosomal ncRNAs are dysregulated and play important roles in different disorders, including several types of cancer, myocardial infarction, and cholestatic liver diseases. Exosomal ncRNAs may be more functional and integral because they are protected from RNase. So exosomes can be suitable places for RNAs crosstalk and an ideal study model of the ceRNA hypothesis.
The ceRNA theory proposes that RNA could regulate each other by competing for miRNA response elements. The ceRNA theory provides a new perspective on the lncRNA regulatory mechanism. Therefore, systematically constructing the EMs-associated lncRNA-miRNA-mRNAs-ceRNA regulatory networks is crucial and might provide more clues to EM molecular mechanisms.
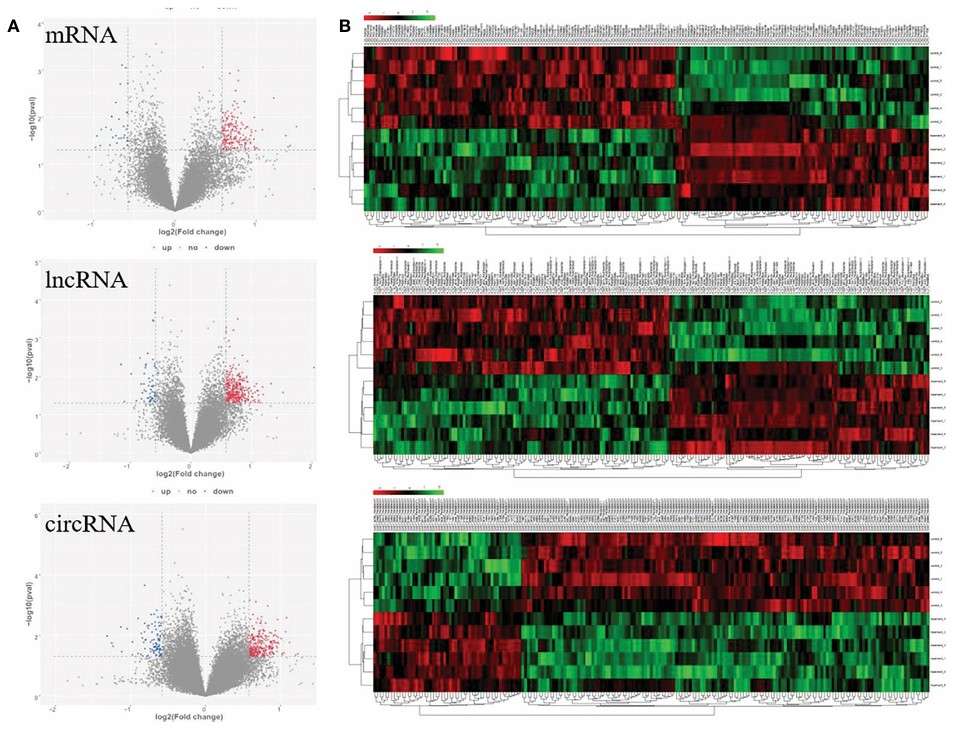 Fig. 2 Volcano plots and clustering heat map for significantly differential genes response to paclitaxel treatment.2
Fig. 2 Volcano plots and clustering heat map for significantly differential genes response to paclitaxel treatment.2
Services at Creative Biolabs
As an industry-leading exosome research services provider, Creative Biolabs launches exosomal ceRNA microarray services to solve our customers' puzzles in exosomal ceRNA. Compared with the traditional NGS method, our exosomal ceRNA microarray requires fewer amount samples and provides higher efficiency.
-
Sample Collection and Isolation
-
Isolate exosomes using ultracentrifugation, precipitation, or other isolation methods.
-
RNA Extraction
-
Extract RNA from isolated exosomes.
-
ceRNA Microarray Analysis
-
Label RNA samples and hybridize them onto ceRNA microarrays.
-
Scan microarrays to detect fluorescence signals indicative of RNA expression levels.
-
Data Analysis
-
Analyze microarray data to identify differentially expressed ceRNAs.
-
Perform bioinformatics analysis to infer ceRNA networks and biological pathways.
Features
-
High Sensitivity and Specificity: Detects low-abundance exosomal RNAs with high fidelity.
-
Comprehensive Profiling: Provides a broad view of ceRNA expression profiles in exosomes.
-
Bioinformatics Support: Includes bioinformatics tools for data interpretation and pathway analysis.
-
Customizable: Option for custom microarray designs tailored to specific research needs.
If you have problems in exosomal ceRNA research, or you have any questions about Creative Biolabs' services, please feel free to contact us for more information.
FAQs
Q: How reliable are the results from exosomal ceRNA microarrays?
A: The microarrays offer high reproducibility and reliability due to stringent quality control measures and optimized protocols.
Q: Can custom panels be designed for specific research questions?
A: Custom microarray panels can be designed based on specific gene targets or pathways of interest.
Q: What are the advantages of studying ceRNAs in exosomes compared to other RNA sources?
A: Exosomal ceRNAs are protected from degradation and reflect the physiological state of cells, making them ideal for biomarker discovery and disease monitoring.
Q: How long does the entire exosomal ceRNA microarray analysis typically take?
A: The timeline can vary, but it generally takes 8-12 weeks from sample collection to data analysis, depending on sample size and complexity.
References
-
Mao, Mingwen, et al. "Role of exosomal competitive endogenous RNA (ceRNA) in diagnosis and treatment of malignant tumors." Bioengineered 13.5 (2022): 12156-12168. Under Open Access license CC BY 4.0, without modification.
-
Lei, Jun, et al. "Analysis of exosomal competing endogenous RNA network response to paclitaxel treatment reveals key genes in advanced gastric cancer." Frontiers in Oncology 12 (2022): 1027748. Under Open Access license CC BY 4.0. The image was modified by revising the title.
For Research Use Only. Cannot be used by patients.
Related Services:

 Fig. 1 The regulatory mechanism of exosomal ceRNA.1
Fig. 1 The regulatory mechanism of exosomal ceRNA.1
 Fig. 2 Volcano plots and clustering heat map for significantly differential genes response to paclitaxel treatment.2
Fig. 2 Volcano plots and clustering heat map for significantly differential genes response to paclitaxel treatment.2









