Overview Service Features Published Data FAQs Scientific Resources Related Services
Creative Biolabs offers academic and industrial researchers services on iPSC generation and applications. Now our experienced scientists at Creative Biolabs are able to use our matured qPCR technology for the analysis of iPSC pluripotency gene expression.
Overview of qPCR Analysis for Pluripotency Markers for iPSC
Introduction of Pluripotency Gene Marker
First demonstrated by Shinya Yamanaka's lab in 2006, the induced pluripotent stem cells (iPSCs) can be generated from different adult somatic cell types by the introduction of four transcription factors (OCT4, SOX2, KLF4, and MYC). With the pluripotency similar to embryonic stem cells (ESCs), iPSCs have shown great potential in drug discovery, disease modeling, and regenerative medicine research. However, the possible low efficiency in pluripotency induction during reprogramming process is the major limitation for the iPSCs generation. In recent years, a number of molecular markers have been identified to verify the pluripotent status of iPSCs which includes Oct4, Nanog, Sox2, Klf4, Ecat, Eras, Fbx15, and Rex1. The process to verify the pluripotency of iPSCs according to well-established markers would be much helpful for the downstream proliferation and differentiation.
Brief Introduction of qPCR and rtPCR
A quantitative polymerase chain reaction (qPCR), also known as real-time polymerase chain reaction (Real-Time PCR) is a commonly used technology of molecular biology based on the polymerase chain reaction (PCR). The amplification of a targeted DNA molecule can be monitored in real-time during the PCR process rather than the end in conventional PCR. In general, there are two methods for the detection of PCR products in qPCR. One is that intercalation of non-specific fluorescent dyes with all the double-stranded DNA. The other is the sequence-specific DNA probes consisting of oligonucleotides that are labeled with a fluorescent reporter which can be detected only after hybridization of the probe with its complementary sequence.
Since pluripotency modification is demonstrated via different protein synthesis and this originates from gene expression result change. As the source of gene expression alteration, mRNA dynamics is the basis and target in discovering biochemical configuration. Since mRNA is single-strained and can not be assessed via quantitative polymerase chain reaction, it is essential in firstly transferring single-strained mRNA into double-strained complementary DNA (cDNA) in a reaction catalyzed by the enzyme reverse transcriptase. The reverse transcriptasing process along with following polymerase chain reaction form the complete process called rtPCR.
rtPCR Analysis for Pluripotency Markers
In order to analyze the pluripotent status of iPSCs, we performed rtPCR for the detection of expressed pluripotency gene markers. Now we are able to use both the human iPSC qPCR kit and mouse iPSC qPCR kit to confirm the iPSC with well-established pluripotency markers.
Mouse Kit includes: NANOG, OCT4, SOX2, TBX3, ESRRB, Tcl1, Actin (positive control).
Human Kit includes: NANOG, OCT4, SOX2, UTF1, hTERT, ZFP42 (REX1), DEMT3B, GAPDH (housekeeping gene).
The Main Protocol
-
Primer design: all the primers used for qPCR should be designed between introns to avoid the amplification of genomic DNA.
-
Quantitative real-time RCR: the final volume is 20μl.
-
Statistical analysis: the qPCR data are from software and compared with the undifferentiated iPSCs.
Creative Biolabs is glad to provide the qPCR and rtPCR analysis for iPSC pluripotency gene markers in custom pathways. Besides,corresponding kits for RNA isolation & purification, RNA transverse transcriptasing and DNA polymerase chain reaction are available for customers' customized operation. Based on our extensive experience, we have the capability to enable you to free up your time for core work and project. If you are interested in our services, please feel free to contact us for more details.
Services at Creative Biolabs
Our qPCR service offers a robust and reliable method for analyzing gene expression levels, making it an instrumental technique for detecting pluripotency markers in iPSCs. Here are the key components of our service.
-
Sample Preparation: We are equipped to handle and process a wide range of sample types, such as tissues, cells, and purified RNA. Our team ensures the integrity of the RNA during the extraction process and we use state-of-art techniques for accurate quantification and qualification. Furthermore, we can also process samples with low RNA quantity or quality with our optimized protocols.
-
cDNA Synthesis: We use a reliable and efficient reverse transcription process for the conversion of RNA into cDNA, which can be used as a template for qPCR analysis. This process allows for the quantification of low-abundant mRNA and non-coding RNA.
-
Primer Design & Validation: Our expert team will design and validate specific qPCR primers for your pluripotency markers. We only use validated primers to ensure high specificity, efficiency, and reproducibility.
-
qPCR Analysis: We utilize advanced qPCR platforms and optimized program protocols to carry out the amplification process, designed to detect and quantify the presence of the specific pluripotency markers in your iPSCs.
-
Data Analysis & Reporting: We provide comprehensive data analysis that includes Ct value determination, efficiency correction, normalization against reference genes, relative quantification, and statistical analysis. The data we provide will be ready for interpretation and publication.
Features of Our Services
-
Accuracy and Precision - Our qPCR technology provides highly accurate and precise quantitation of the specific pluripotency markers expressed in iPSCs. This can help in validating the pluripotency status of the generated iPSCs more convincingly and ensures the reliability and reproducibility of your results.
-
Speed and Efficiency - Our qPCR services offer rapid turn-around time.
-
Comprehensive Analysis - Our service includes full analyses of pluripotency and differentiation genes, ensuring you have a comprehensive understanding of the iPSC's status. This not only checks the expression of pluripotency genes but also rules out differentiation along any of the germ layers.
-
Cost-Effective - We can reduce the need for expensive lab equipment and specialized staff in your own establishment.
Published Data
Below are the findings presented in the article related to qPCR analysis for pluripotency markers for iPSC.
Valentina, et al. characterized iPSC clones comparatively using morphological appearance, alkaline phosphatase (AP), immunocytochemistry, protein blotting, and RT-qPCR analyses in order to test whether changes in reprogramming capacity affect the pluripotent properties of the generated iPSC clones.
The results showed that the expression levels of endogenous pluripotent genes (OCT4, SOX2 and NANOG) were comparable in all characterized clones as well as in hESC and hiPSC cell lines. In contrast, fibroblasts used as negative controls did not show any expression of these genes. This data suggests that all generated iPSC clones have the same morphological appearance, biochemical activity, and known pluripotent gene expression, regardless of their phenotype.
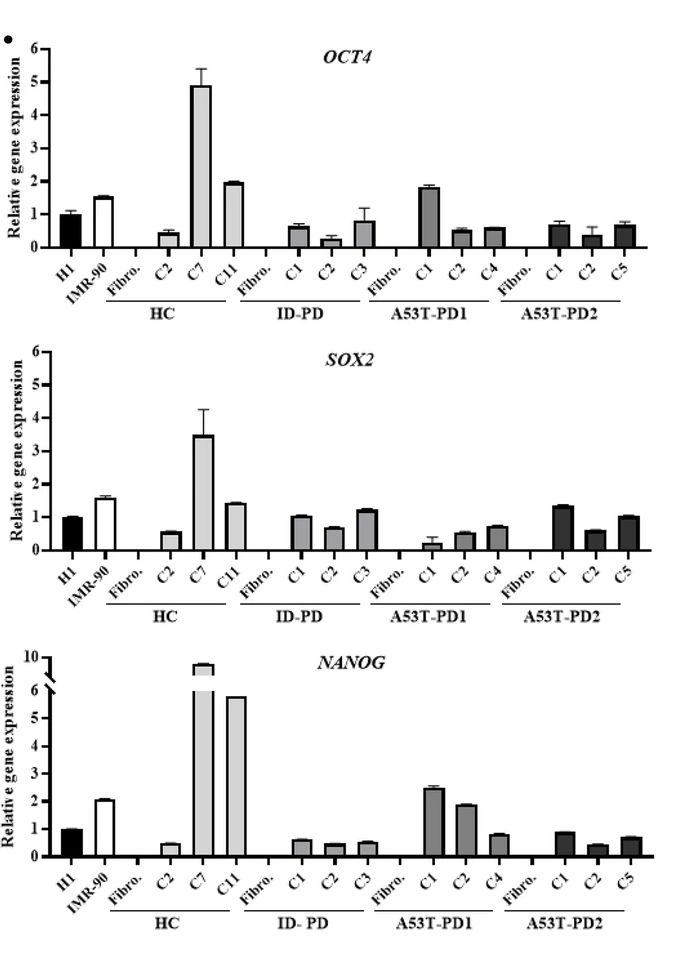 Fig. 1 RT-qPCR analysis of pluripotent genes expressed by the generated iPSCs.1
Fig. 1 RT-qPCR analysis of pluripotent genes expressed by the generated iPSCs.1
FAQs
-
Q: What markers are typically analyzed in this qPCR service?
A: In our qPCR analysis service, we typically analyze a panel of core pluripotency markers, including OCT4 (POU5F1), SOX2, NANOG, LIN28, KLF4, and c-MYC. These markers are well-established indicators of the pluripotent state and are commonly used to assess the quality and characteristics of iPSCs.
-
Q: How much of my iPSC sample do you need for the qPCR analysis?
A: For our qPCR analysis, we generally require a small amount of RNA extracted from your iPSC samples, usually around 1-2 micrograms. This quantity ensures that we have sufficient material to perform the analysis accurately and provide reliable results. If you have concerns about the sample size, please let us know, and we can discuss the specific requirements based on your sample's characteristics.
-
Q: How do you ensure that the qPCR analysis is accurate and reliable?
A: We ensure the accuracy and reliability of our qPCR analysis through several measures: using high-quality reagents and standards, following stringent protocols, performing technical replicates, and including positive and negative controls in each run. Our lab is equipped with advanced qPCR instruments and our technicians are highly trained, ensuring consistent and precise results.
-
Q: What happens if my iPSCs do not express the expected pluripotency markers?
A: If your iPSCs do not express the expected pluripotency markers, our report will highlight this discrepancy and provide potential reasons, such as issues with reprogramming efficiency, cell culture conditions, or sample handling. We can offer troubleshooting advice and recommend additional tests or steps to improve the quality of your iPSCs. Ensuring the expression of these markers is vital to the success of your research and applications.
Scientific Resources
Reference
-
Swaidan, Nuha T., et al. "Identification of potential transcription factors that enhance human iPSC generation." Scientific Reports 10.1 (2020): 21950. Distributed under Open Access license CC BY 4.0. The image was modified by extracting and using only Part C of the original image.
For
Research Use Only. Not For Clinical Use.
 Fig. 1 RT-qPCR analysis of pluripotent genes expressed by the generated iPSCs.1
Fig. 1 RT-qPCR analysis of pluripotent genes expressed by the generated iPSCs.1