Creative Biolabs has long-term devoted to the generation and application of iPSCs. Based on years of experience, now we can provide the pluripotency marker analysis for iPSC enrichment and isolation. Furthermore, we can also provide a customized proposal to meet your project requirements.
With the similar properties with human embryonic stem cells (hESCs) both at molecular and functional levels, human induced pluripotent stem cells (hiPSCs) are considered to be potential for drug screening, disease modeling, and regenerative medicine. Now, hiPSCs can be generated from different somatic cell types by overexpressing transcription factor combination. However, there is a major limitation during reprogramming process that low efficiency in pluripotency induction leads to the generation of a few reprogrammed hiPSCs. Although the morphology analysis of iPSCs has been used for their isolation, some studies have shown that the isolated hiPSC clones have differences in the expression of pluripotency gene markers and those with the similar levels of pluripotency marker expression present different abilities in lineage differentiation. In this case, the analysis of pluripotency gene marker expression would be a potential strategy for the enrichment and isolation of iPSCs.
After a series of studies, the expression of three pluripotency markers, SSEA-4, TRA-1-60, and NANOG, have been analyzed by immunofluorescence staining in emerging iPSCs. We have found that the reprogramming cells achieve the SSEA-4+TRA-1-60+NANOG+ pluripotent state in the very early days of reprogramming and the number of this kind of cells increases with the colony size increased. SSEA-4+ cells are molecularly heterogeneous throughout reprogramming according to the temporal changes whereas the TRA-1-60+ cells are less heterogeneous. Different markers present function signal in different stages. The emergence of SSEA-4 signal occurs in the early stages, while TRA-1-60 signal appears in the intermediate stages and the expression of NANOG is a definitive marker of pluripotency. In summary, the use of established markers to verify stem cell pluripotency at the start of an experiment is a good method to ensure the pluripotency states for the studies of iPSCs proliferation and differentiation.
 Fig.1 Expression of the pluripotency markers (SSEA-4, TRA-1-60, and NANOG) in the colonies during reprogramming.1
Fig.1 Expression of the pluripotency markers (SSEA-4, TRA-1-60, and NANOG) in the colonies during reprogramming.1
Apart from the established markers, we are looking for the other pluripotency gene markers for iPSCs. On the basis of your special requirement, we can help for iPSCs culture, reprogramming, and differentiation. Based on our mature flow cytometry and immunofluorescence technologies, we provide high-efficient pluripotency marker assays for the enrichment and isolation of iPSCs. Creative Biolabs also provides other iPSC-related services including the generation and applications, please feel free to contact us if you are interested in our services.
Our pluripotency marker assays for iPSCs provide comprehensive analysis to confirm the pluripotent state of iPSCs. This service is designed for research use only and is tailored to meet the rigorous standards required in stem cell research. Our assays offer a robust suite of tests to verify the expression of key pluripotency markers at both the transcriptional and protein levels.
Our specific workflow includes:
By choosing our pluripotency marker assays, researchers can confidently validate the pluripotent state of their iPSC lines, ensuring the reliability and robustness of their downstream applications.
Below are the findings presented in the article related to pluripotency marker assays for iPSC.
Carol X.-Q. Chen, et al. established a multi-step workflow to evaluate newly generated iPSCs. The workflow tested four benchmarks: cell growth, genomic stability, pluripotency, and the ability to form three germline layers. The workflow is scalable and specialized for use with iPSC for research applications.
In their workflow, each iPSC cell line was examined for pluripotency. As detected by immunohistochemistry (ICC), all tested iPSC cell lines expressed the pluripotency markers SSEA-4, OCT3/4, NANOG, and Tra-1-60R, which were the same as the H9 ESC cell line. There was no difference in the fluorescence intensity of these markers in these cell lines. These iPSCs were not only morphologically similar to each other and to ESCs, but also to many common pluripotency markers (OCT3/4, SOX2, NANOG) at the transcriptional level.
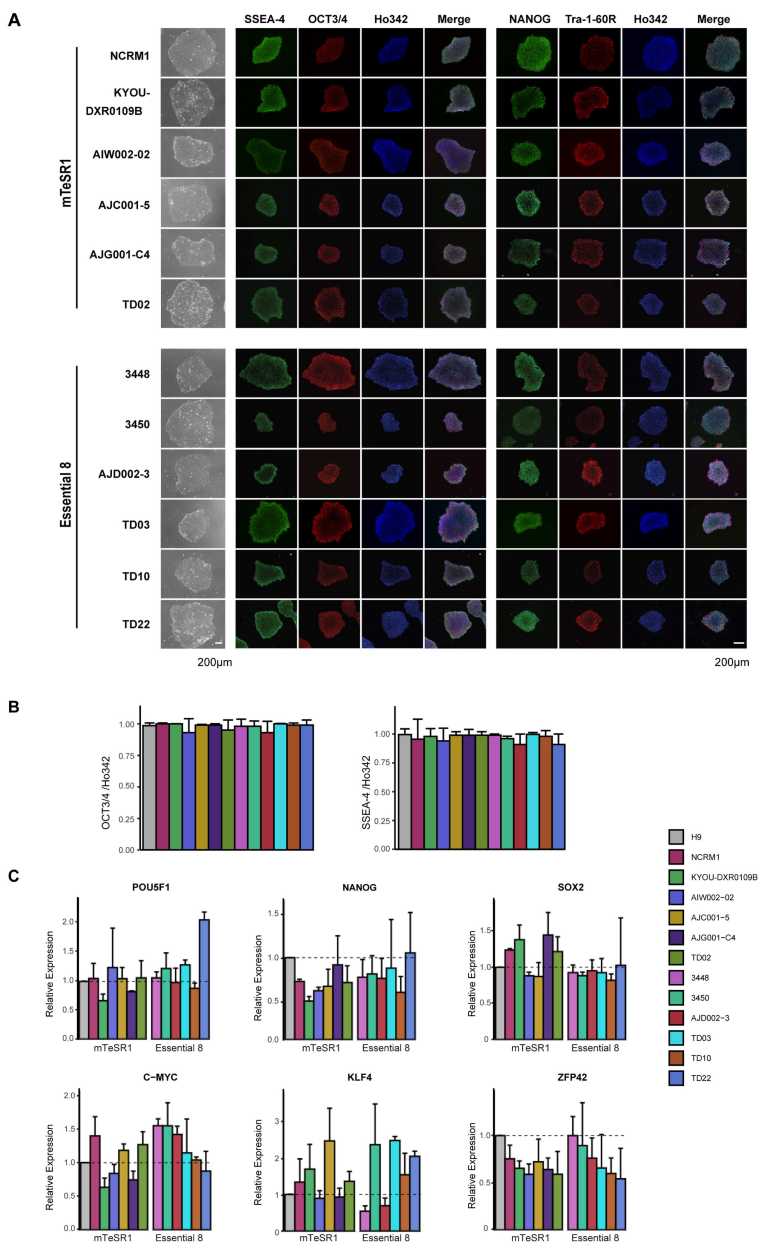 Fig. 2 Expression of pluripotency markers in hiPSCs.2
Fig. 2 Expression of pluripotency markers in hiPSCs.2
References
For Research Use Only. Not For Clinical Use.