Identification of B-cell epitopes is a fundamental step for the development of epitope-based vaccines, therapeutic antibodies, and diagnostic tools. Creative Biolabs offers a comprehensive range of B-cell epitopes mapping services to accelerate the customer's project progress.
Antigen-antibody interaction is a key event in the humoral immune response. In this process, specific antibodies recognize antigens at discrete regions known as antigenic determinants or B-cell epitopes. B-cell epitopes can be defined as a surface accessible clusters of amino acids, which are recognized by secreted antibodies or B-cell receptors and are capable of eliciting a cellular or humoral immune response. Based on the spatial structure of the B-cell epitopes, it can be divided into two groups: linear (continuous or sequential) and conformational (nonlinear or discontinuous) epitopes. Linear epitopes are formed by linear stretch residues in the antigenic protein sequence and typically vary from 5 to 20 amino acids in length. In contrast, conformational epitopes are formed by residues that are far apart in the antigen sequence, and these residues are brought together in space by their folding. Although the majority of epitopes are conformational, most of them are composed of 1-5 linear stretches. Linear B-cell epitope has vast application in the area of antibody production, immunodiagnostics; epitope-based vaccine design, selective deimmunization of therapeutic proteins and autoimmunity.
 Fig.1 B cell epitope.
Fig.1 B cell epitope.
B cell epitope identification is essential for the development of vaccines and the selection of high-affinity antibodies for immunotherapy and immunodiagnostics. Currently, some reliable B cell epitope mapping tools have been applied to many clinical and biotechnology research, such as vaccine design and therapeutic antibody development. Experimental methods developed to identify epitopes can be broadly classified into structural and functional studies.
Structural approaches
The most accurate method for structural epitope mapping is X-ray crystallography of antigen-antibody (Ag-Ab) complexes, and this method is generally considered to be the only way to define structural epitopes and is a guarantee for the precise identification of linear and conformational epitopes. Bacterial or viral antigens, especially small soluble proteins, are ideal for crystallography. However, there are some limitations in the application of X-ray crystallography, such as being limited by the quality of cocrystals and electron density of the antibody. In recent years, other methods, including nuclear magnetic resonance (NMR) and electron microscopy (EM), have been used to replace traditional X-ray crystallography. NMR shows several advantages over X-ray crystallography, such as providing data on the structure, kinetics, and binding energy of the Ag-Ab complex, and being able to perform in solutions where no crystals are needed. It should be noted hat NMR is limited to small proteins and peptides (<25 kDa). EM can also be used for epitope mapping, and it is a low-resolution structural approach for larger antigens (e.g., whole viral particles).
Functional approaches
Most B cell epitope mapping experiments make use of linear peptide fragments from antigenic proteins. These peptides can be homologous enough to parts of the whole antigen to allow for antibody binding. Some of the commonly used methods for functional B-cell epitope mapping are screening of antigen-derived proteolytic fragments or peptides for antibody binding and testing the Ag-Ab reactivity of mutants (site-directed or randomly mutated). Other techniques, such as display technologies and mimotope analysis have also become acceptable alternative choices for epitope mapping because of their relative cost, flexibility, and speed.
Creative Biolabs' experts can provide customized solutions according to your project needs. Our unique solutions for immunology research can save you time, money and risk, whether your objective is basic research, preclinical or clinical development. Please contact us for more details.
Other optional CreMap™ B cell epitope mapping services:
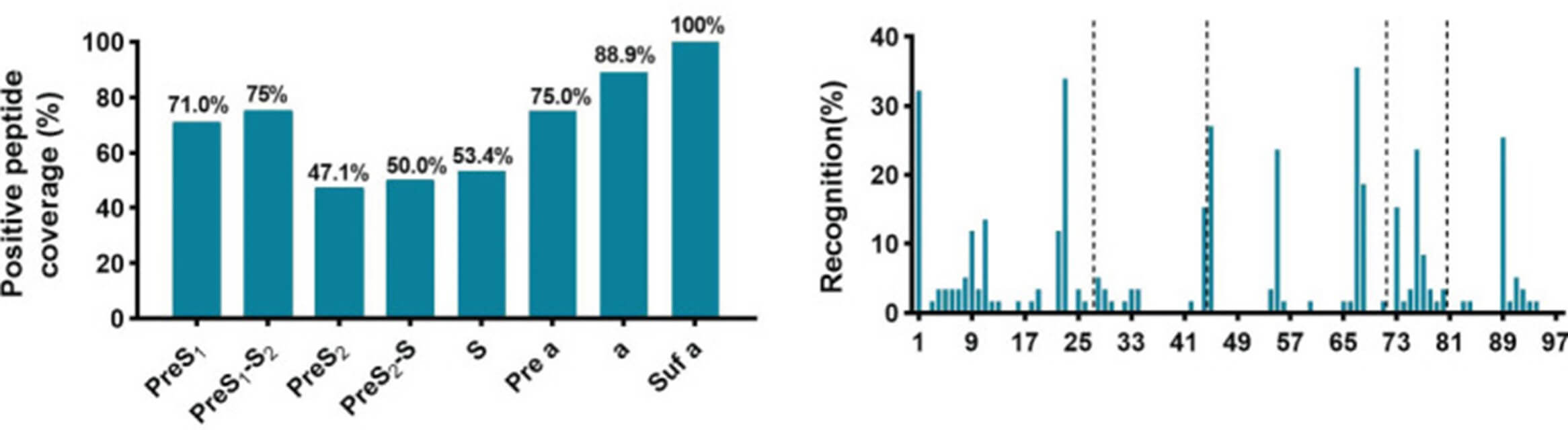 Fig. 2 General recognition of linear B-cell epitopes on S protein by peptide array. (Shuqin Gu, 2021)
Fig. 2 General recognition of linear B-cell epitopes on S protein by peptide array. (Shuqin Gu, 2021)
The study focuses on identifying B-cell linear epitopes associated with hepatitis B surface antigen (HBsAg) loss in patients treated for chronic hepatitis B virus (HBV) infection, using a peptide array. The significance lies in pinpointing specific epitopes that correlate with a favorable immune response and potential clinical remission. Linear B cell epitope mapping was employed using a peptide array that systematically tested overlapping peptides from HBV proteins against sera from different patient groups. This method successfully identified several dominant linear epitopes that were frequently recognized by sera from patients who achieved HBsAg loss, suggesting these epitopes as targets for therapeutic vaccine development aimed at eliciting robust neutralizing antibodies.
Linear B cell epitope mapping is a technique used to identify the specific linear sequences of amino acids in an antigen that are recognized by B cell antibodies. Unlike conformational epitopes, which depend on the protein's three-dimensional structure, linear epitopes consist of continuous amino acid sequences. This mapping is crucial for understanding immune responses and designing vaccines and therapeutic antibodies.
Linear B cell epitope mapping involves synthesizing overlapping peptides that span the entire sequence of an antigen. These peptides are then tested for their ability to bind to antibodies using assays such as ELISA or microarray. By identifying which peptides are recognized by the antibodies, researchers can determine the specific linear sequences that constitute the epitopes.
Linear B cell epitope mapping has simplicity and the ability to identify epitopes without requiring the antigen's three-dimensional structure. This method allows for the high-throughput screening of multiple peptides, making it efficient and cost-effective. It also provides detailed information about the exact amino acid sequences recognized by antibodies, which is valuable for vaccine and therapeutic development.
Linear B cell epitope mapping is beneficial for vaccine development, therapeutic antibody design, and diagnostic tool creation. It helps identify immunogenic regions of pathogens, understand antibody specificity, and develop targeted therapies. This technique is also valuable in studying autoimmune diseases, allergy responses, and developing personalized medicine approaches based on specific epitope profiles.
Linear B cell epitope mapping contributes to vaccine development by identifying specific sequences within antigens that elicit strong antibody responses. By targeting these linear epitopes, researchers can design vaccines that effectively stimulate the immune system. This approach helps in creating vaccines with higher efficacy and safety profiles by focusing on the most immunogenic and protective regions of the pathogen.
Linear B cell epitope mapping is limited to identifying continuous amino acid sequences and cannot detect conformational epitopes, which are formed by discontinuous sequences brought together by the protein's three-dimensional structure. This limitation means that some critical epitopes may be missed. Additionally, linear epitope mapping may not fully capture the complexity of antibody-antigen interactions in their native conformations.
Linear B cell epitope mapping can be applied to virtually any antigen, as long as the amino acid sequence of the antigen is known. This versatility makes it suitable for studying a wide range of pathogens, including viruses, bacteria, and parasites, as well as for identifying epitopes in cancer and autoimmune diseases. The technique is adaptable to various research needs and applications.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
