Based on enriched experience and professional technical team, Creative Biolabs provides Alanine Scanning Mutagenesis library to help our clients performing B cell epitope mapping. And this method could identify the crucial amino acid residues of certain target peptides which contribute to the peptide’s functionality.
 Fig 1. Alanine scanning library.
Fig 1. Alanine scanning library.
Alanine scanning mutagenesis technology is a useful and powerful approach to construct the peptide library (alanine scanning library) for identifying the B cell epitopes. Because of its inert methyl functional group, Alanine is often used on our platform to characterize the contribution of a specific amino-acid. Libraries involving other mutagenesis are also available when required. Each non-alanine residue of the target peptide will be sequentially substituted by one alanine at a time, which we could construct the mutation peptide library. A highly-specified site-directed mutagenesis is introduced in this crucial step. Then hundreds of plasmid clones from the mutation library will be expressed in mammalian cells. Subsequently, the corresponding change of the peptide-antibody binding activity will be measured. The antigen whose key amino-acid is replaced by the alanine loses or changes the binding ability to the specific antibody. Substitution of the key amino acid residue with alanine will lead to the mutation peptide’s failure of binding to antibodies or changes in the binding activity. Substitution with alanine residues eliminates side-chain interactions without altering the backbone conformation or introducing steric or electrostatic effects, so it is often the preferred choice for testing the contribution of specific side-chains while preserving native protein structure.
CreMap™ alanine scanning mutagenesis mapping platform is a high-throughput strategy for mapping both linear and conformational antigen epitopes by evaluating the effects of point mutations across a target protein. And customs could obtain precise information about the functional binding sites of the target peptide at the level of amino acid. If you are interested in this service, please contact us for more information.
Other optional CreMap™ B cell epitope mapping services:
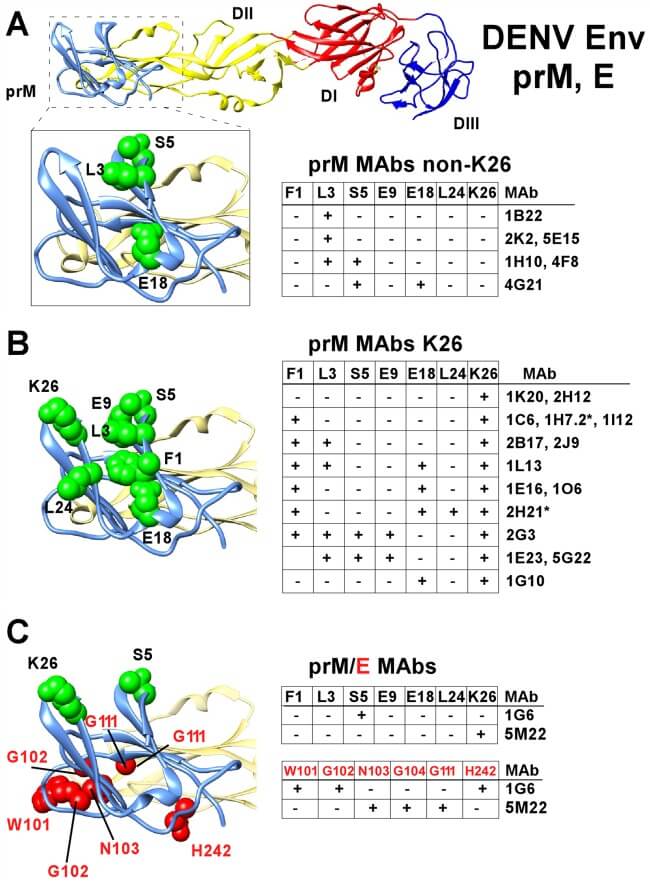 Fig. 2 Epitope mapping of prM region antibodies by shotgun mutagenesis library screening. (Scott A. Smith, 2016)
Fig. 2 Epitope mapping of prM region antibodies by shotgun mutagenesis library screening. (Scott A. Smith, 2016)
The research explored the interactions between human monoclonal antibodies and the dengue virus premembrane (prM) protein, focusing on those with virus replication-enhancing properties. The study identified a single immunodominant antigenic site on the prM protein recognized by all isolated antibodies. Alanine scanning mutagenesis was used to pinpoint specific residues within this epitope that are crucial for antibody binding. This technique involved systematically substituting alanine at different positions within the prM protein and observing the impact on antibody binding, helping to delineate the fine specificity of antibody interactions. Such detailed mapping is critical for understanding how dengue virus evades the immune system and enhances its replication, which is pivotal for developing more effective vaccines and therapies against dengue.
Alanine scanning mutagenesis is a technique used to identify the specific amino acids within a protein that are critical for binding interactions, such as those between an antigen and an antibody. By systematically substituting each amino acid in the protein with alanine, researchers can determine which residues are essential for binding.
In alanine scanning mutagenesis, each amino acid in the epitope region of a protein is individually replaced with alanine, a small, non-reactive amino acid. The impact of each substitution on the protein's ability to bind to its partner (like an antibody) is then measured. This process reveals which amino acids are crucial for the interaction.
Alanine scanning provides precise information about the functional role of each residue within a protein interaction interface. It allows researchers to pinpoint which amino acids are crucial for binding, thereby identifying key areas of the protein that could be targets for drug development or vaccine design.
Alanine scanning mutagenesis is versatile and can be applied to virtually any protein. However, its effectiveness may be limited for proteins where the replacement of residues with alanine leads to misfolding or loss of structural integrity, which could obscure true binding interactions.
While traditionally more labor-intensive and low-throughput, advances in molecular biology techniques and automation have enabled high-throughput alanine scanning studies. These involve parallel processing of many mutations and can significantly accelerate the mapping process.
Unlike methods that require visualizing the physical structure of protein complexes, like X-ray crystallography, alanine scanning offers a functional perspective by directly testing the impact of specific amino acid changes on binding. It is often used in complement to structural techniques to validate and extend structural data.
While effective at identifying critical residues, alanine scanning does not provide information about the overall structure of the protein or complex. It also does not indicate whether the modifications may alter the protein's function in ways not directly related to binding the target molecule.
Alanine scanning can be used to analyze multiple epitopes on a single antigen. However, the approach requires careful design to isolate the effects of mutations within each epitope, especially when epitopes are close together or overlap structurally.
The outcome of an alanine scanning experiment typically includes a map of essential residues within the epitope, highlighting which amino acids are indispensable for binding. This map is crucial for understanding the molecular basis of the interaction and for guiding further modifications to enhance binding or alter the immunogenic properties of the protein.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
