Creative Biolabs provides a versatile genetic tool MAPPIT that can be used to analyze and detect new protein-protein interactions in mammalian cells. Discerning the interacting partners of a protein is a straightforward method to gain insight into the protein's functionality and to position it in an interaction network within cells. Varieties of biotechnologies have been described to serve this purpose, and some of them are specially used to study posttranslational modifications in mammalian proteins so as to clarify their intrinsic physiological context. Whereas, several inherent constraints restrict the use of these techniques and most are not suitable for discovering new interacting partners. It’s worth noting that a novel protocol termed Mammalian Protein-Protein Interaction Trap (MAPPIT) can work as a versatile genetic tool to detect and analyze unknown protein-protein interactions in mammalian cells.
 Fig.1 Overview of the different MAPPIT approaches.
Fig.1 Overview of the different MAPPIT approaches.
(a) MAPPIT (b) Heteromeric MAPPIT (c) Reverse MAPPIT (d) MASPIT (Mammalian small molecule-protein interaction trap)
Physical interactions between proteins play an important role in cellular processes. Focus on drawing the protein interaction maps is ongoing in a number of model organisms and will provide a scaffold for further detailed functional research by a diversity of approaches. MAPPIT is a mammalian two-hybrid system that allows the detection and analysis of protein-protein interactions (PPI), particularly in their native environment. This method is on the basis of type I cytokine receptor signal transduction. A bait protein of interest is fused to the signaling-deficient cytokine receptor chimera, the signaling competence of which is restored upon recruitment of a prey protein coupled to a functional cytokine receptor domain. Ultimately, the result leads to the transcription of a reporter or marker gene under the control of the promoter.
MAPPIT stands out as a pioneering PPI technology which is found on a true mammalian signal transduction pathway, the Janus kinase-signal transducer and activator of transcription (JAK-STAT) cascade, other than derived from yeast or other lower animal models. MAPPIT has arisen from the knowledge of the classic JAK-STAT pathway. In mammals, there are four JAK kinases (JAK1, JAK2, JAK3, and Tyk2) and seven STAT transcription factors (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6) having been identified. The canonical pattern of JAK-STAT signaling is activated by the binding of specific peptide ligands (e.g. cytokines as interleukins and colony-stimulating factors, several hormones as leptin and growth hormone) to transmembrane receptors.
The MAPPIT assay typically utilizes a signaling-deficient chimeric type I cytokine receptor whose extracellular domain is derived from the homodimeric erythropoietin receptor (EpoR) and is fused to the transmembrane domain and cytoplasmic tail of the leptin receptor (LepR). This chimeric receptor causes signaling-deficient by mutating the three tyrosines (Tyr) presented in the cytoplasmic tail to phenylalanine (Phe). Through ligand administration, the associated JAKs of this chimeric receptor is activated. However, since there is no tyrosine left in the receptor tail to be phosphorylated, no STAT recruitment sites will be initiated and no signal is transmitted to the nucleus as well. The protein of interest that will serve as bait is C-terminally fused to this signaling-deficient receptor. The prey protein referred to test for interaction with the bait one is coupled to a portion of another cytokine receptor, the glycoprotein 130 receptor (gp130). The usage of gp130 domain contains Tyr motifs after phosphorylation via the JAKs that work as STAT3 recruitment sites. If bait and prey, two proteins of interest interact, the JAKs will be able to phosphorylate Tyrs with cytokine ligand activation. That result generates functional STAT3 binding sites and recruited STATs upon activation by JAKs will migrate to the nucleus to activate the transcription of a luciferase reporter gene.
 Fig.2 Schematic representation of the MAPPIT principle.
Fig.2 Schematic representation of the MAPPIT principle.
The interaction between bait and prey results in the recruitment of a gp130 fragment containing STAT3 recruitment sites and thereby facilitating the complementation of the signaling-deficient chimeric receptor bait. In a nutshell, MAPPIT depends on a dysfunctional JAK-STAT pathway, of which the activity is only restored when PPI of specific bait and prey occurs.
Creative Biolabs has seasoned technicians to help worldwide scientists to detect designated protein-protein interactions for their research. MAPPIT system has been proven to be valuable to study interactions between proteins in mammalian cells where offer a normal physiological context for mammalian proteins to be tested. This highlighted asset ensures proper folding and provides the necessary cofactors and regulatory proteins participating in post-translational modifications or assisting any conformational alterations to achieve protein’s interaction.
Other strengths of MAPPIT technique include being sensitivity, robustness, and scalability. It exhibits a significant signal-to-noise ratio, identifies various protein interactions including transient and indirect ones, and has been shown to be highly complementary to other two-hybrid assays. Since the conception of the original MAPPIT, the platform has been expanded assay variations and permitted large-scale PPI screening, mapping of PPI interfaces, PPI inhibitor screening, and drug profiling which open up new fields of application. Meanwhile, there are additional tools of MAPPIT aiming at increasing the versatility of that platform.
 Fig.3 The MAPPIT toolbox and its applications.
Fig.3 The MAPPIT toolbox and its applications.
Other optional protein-protein interaction (PPI) assay services:
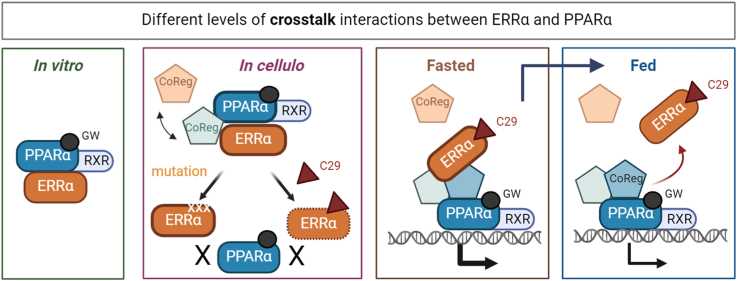 Fig. 4 Different levels of crosstalk interactions between ERRα and PPARα. (Sofie J. Desmet, 2024)
Fig. 4 Different levels of crosstalk interactions between ERRα and PPARα. (Sofie J. Desmet, 2024)
Peroxisome proliferator-activated receptor α (PPARα) is a transcription factor that can drive target genes involved in fatty acid β-oxidation. The ORFeome range unbiased mammalian protein-protein interaction trap (MAPPIT) using PPARα as bait revealed that PPARα-ligand-dependent interaction and orphan nuclear receptor estrogen-related receptor α (ERRα). The researchers used orthogonal protein-protein interaction analysis and ERRα pharmacological inhibitors to confirm their functional interactions and to study the effects of crosstalk mechanisms. Random mutagenesis screening and structural superposition are used to characterize the interaction surface and contact point. Through coregulatory peptide recruitment analysis, the scientists determined the degree of mutual ligand effect between the two nuclear receptors. ERRα chromatin immunoprecipitation analysis was performed in the liver of fasting and eating mice, and the researchers revealed the PPARα target from genome-wide transcriptome analysis. The results show that there is a multi-layer transcriptional crosstalk mechanism between PPARα and ERRα, which may help to fine-tune the activity of PPARα as a nutrition-induced transcription factor.
The mammalian protein-protein interaction trap (MAPPIT) is a technique used to study protein-protein interactions within mammalian cells. This method relies on a modified cytokine receptor that can signal through a pathway only when a specific protein-protein interaction occurs.
MAPPIT allows the study of protein-protein interactions within the context of a living mammalian cell, providing insights that are more physiologically relevant compared to in vitro assays. The technique provides a quantitative readout of interactions through reporter gene activation, enabling the assessment of interaction strength and dynamics. MAPPIT can be adapted to study various types of interactions by modifying the bait and prey proteins, making it a versatile tool for different research applications. The method allows for real-time monitoring of interactions, which can be particularly useful for studying dynamic processes and temporal changes in protein interactions.
The mammalian protein-protein interaction trap (MAPPIT) can be effectively used to identify new protein-protein interactions through a systematic screening approach. Here's a step-by-step outline of the process:
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
