Protein-protein interactions are closely associated with many biological processes, such as transcription, DNA modifications, and transcriptional regulations. Exploring the interactions between macromolecules contributes to revealing the molecular mechanisms of life processes. At Creative Biolabs, our experienced team of experts has built a reputation for providing cost-effective yeast three-hybrid (Y3H) services with fast turnaround times. Our comprehensive services range from initial experimental design to final analysis of results, helping you make breakthroughs in complex protein interactions.
Yeast hybrid systems are widely used due to their convenience and low cost. Based on these systems, many methods have been developed to analyze protein-protein, protein-DNA, and protein-RNA interactions. For example, the yeast two-hybrid system (Y2H) plays a crucial role in studying protein-protein interactions. Y3H, an extension of the Y2H, is a powerful tool for studying more complex macromolecular interactions involving three components.
Y2H system is a promising tool for detecting protein interactions. In addition to the case of two proteins interacting, it is also common for a third protein to stabilize the binding between the two partners. The Y3H system has gradually emerged as an attractive strategy to explore such complex protein complexes by introducing a new bait vector, pBridge, which allows to insertion of two exogenous proteins at the same time. In this system, a bait protein is expressed with a DNA binding domain (BD) and then the intermediate protein, and the prey protein is fused with the transcriptional activation domain (AD). The Pmet25 promoter in the pBridge vector only can be expressed in the medium lacking methionine. Thus, the interaction between bait protein and prey protein through the intermediate protein is identified in the medium lacking methionine after BD and AD vectors co-transformed into the yeast strain.
 Fig.1 The yeast three-hybrid system vectors. (Sandrock, 2001)
Fig.1 The yeast three-hybrid system vectors. (Sandrock, 2001)
Similar to the Y2H system, the Y3H system can be applied in various research fields.
As a global company that places great emphasis on the fast delivery of solutions and cost-effective service, Creative Biolabs is committed to providing customized Y3H services for your specific project needs. Please contact us to tell us your needs, we will offer you flexible and innovative solutions and the highest quality Y3H services to help you solve the problems you encounter during your research.
Other optional protein-protein interaction (PPI) assay services:
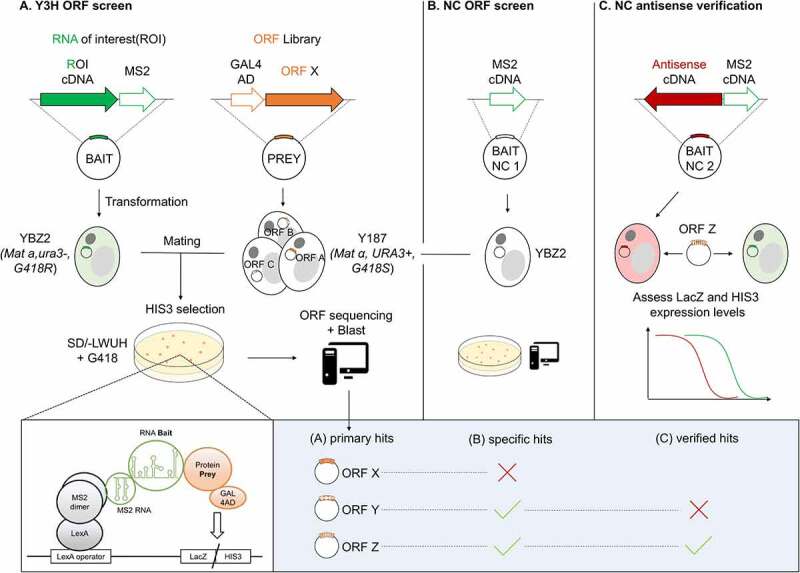 Fig. 2 Detecting RNA-binding proteins in a mating yeast three-hybrid ORF screen. (Sander Jansen, 2021)
Fig. 2 Detecting RNA-binding proteins in a mating yeast three-hybrid ORF screen. (Sander Jansen, 2021)
In the process of infection, extracellular ribonuclease XRN1 can not completely degrade the RNA genomes of dengue virus (DENV) and Zika virus (ZIKV), which will lead to the formation of small subgenomic flavivirus RNA (sfRNA) in infected host cells. These non-coding RNAs are key virulence factors, which always seem to affect the molecular interaction with RNA-binding proteins (RBP) in the cellular process. Here, the researchers used the yeast three-hybrid (Y3H) system for correlation analysis. They improved the RNA-Y3H method to expand the proteome screening of RBP. Through the optimized Y3H system, they screened human proteins bound to DENV and ZIKV sfRNA and obtained a list of 69 presumptive sfRNA conjugates, including several previously reported conjugates and many novel RBP host factors.
The yeast three-hybrid (Y3H) system is an adaptation of the yeast two-hybrid system that is designed to detect and analyze interactions between two proteins and a third molecule, typically an RNA or a small molecule. In this system, one protein is fused to a DNA-binding domain (DBD) and the other to an activation domain (AD). The third component, such as RNA, is linked to a molecule that can bridge the DBD and AD fusions when bound by the two proteins. If all three components interact successfully, the reconstituted DBD and AD drive the transcription of a reporter gene, indicating a positive interaction.
The Y3H system is particularly useful for studying the interactions that involve a third component, such as RNA molecules or small biochemical molecules. This system allows researchers to:
The yeast three-hybrid (Y3H) system is an extension of the yeast two-hybrid (Y2H) system, which is traditionally used to study protein-protein interactions. The Y3H system incorporates a third component, typically an RNA molecule or a small chemical compound, allowing for the investigation of the interaction between two proteins in the presence of this third molecule. This additional component can be critical for interactions that are dependent on or regulated by small molecules or RNA, providing a more nuanced understanding of protein dynamics and function within the cell.
Yes, the yeast three-hybrid system can be adapted to study a wide range of interactions involving various biomolecules, including DNA, peptides, and even certain post-translational modifications. By modifying the third component to include these different molecules, researchers can explore how these elements influence protein interactions. This flexibility makes the Y3H system a versatile tool for studying complex biochemical pathways and molecular interactions that are critical to cellular function and disease processes.
Commonly used reporter genes include HIS3, lacZ, and ADE2.
By quantifying the expression level of the reporter gene, researchers can gauge the strength and stability of these interactions between proteins and the third component under different conditions. This quantitative approach can be enhanced by using fluorescent or luminescent reporters, which allow for real-time, dynamic measurement of interaction strengths. Such quantitative data are invaluable for understanding the kinetics and binding affinities involved in molecular interactions, aiding in detailed mechanistic studies and the design of therapeutic interventions.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
