Creative Biolabs provides a novel technique, proximity-dependent biotin identification approach, recently described as BioID, for screening and identification of protein-protein interactions in vivo. It takes advantage of the ability of a mutant form of biotin ligase to promiscuously biotinylate proximal proteins, no matter directly interacting or vicinal ones.
BioID is a unique and versatile technique designed to detect protein-protein interactions within intact cells. This method employs a promiscuous biotin protein ligase fused to a bait protein, enabling the biotinylation of nearby endogenous proteins during a defined labeling period. Biotinylation occurs regardless of whether these proteins interact directly or indirectly with the fusion protein or are merely located within the same subcellular neighborhood. The biotin-tagged proteins are subsequently captured using streptavidin affinity methods and identified by mass spectrometry. BioID has proven effective across diverse species and cell types, including mammalian cells and cellular structures like the insoluble nuclear lamina and centrosome. This approach is especially valuable for studying insoluble proteins, identifying weak or transient interactions, and regulating interactions over time.
The BioID method draws inspiration from DamID, which uses a prokaryotic Dam methylase to explore DNA-protein interactions in eukaryotic cells. In BioID, the mutated Escherichia coli biotin ligase BirA* serves as the central enzyme, derived from the wild-type BirA (with an R118G mutation). Biotinylation proceeds through a two-step process, where the biotin ligase generates a highly reactive biotinoyl-5'-AMP intermediate that reacts with lysine residues on target proteins, releasing AMP. Unlike the wild-type BirA, which has strict substrate specificity, the BirA* mutation results in promiscuous biotinylation, modifying primary amines in the immediate vicinity of the tagged protein. Due to the short half-life of the intermediate, the labeling radius in BioID is limited to approximately ~10 nm, allowing precise mapping of protein interactions within cellular environments.
 Fig. 1 BioID. The protein of interest is fused with a promiscuous form (BirA*) of the bacterial biotin ligase BirA and expressed in cells.1,5
Fig. 1 BioID. The protein of interest is fused with a promiscuous form (BirA*) of the bacterial biotin ligase BirA and expressed in cells.1,5
There are two main stages splitting out in a BioID process: (i) generate a BioID fusion protein vector for stable expression within a mammalian cell or desired cell line; (ii) utilize transient transfection or the stable cell line to proceed with a large-scale BioID pull-down for validation of potential protein-interactors by mass spectrometry. The overall scheme of BioID steps in detail as follows:
 Fig.2 Overview of proximity biotinylation by BirA*.2,5
Fig.2 Overview of proximity biotinylation by BirA*.2,5
BioID provides multiple advantages over traditional approaches for studying protein-protein interactions.
| BioID | Traditional Techniques | |
| Detection of Weak and Transient Interactions | Capable of identifying weak and transient interactions in vivo | Limited in detecting weak or transient interactions |
| Addressing Solubility Issues | Suitable for studying insoluble or inaccessible protein structures | Often limited by solubility and accessibility issues |
| Elimination of Non-Specific Bindings | Strong biotin-streptavidin binding allows harsh washing conditions | Less tolerant to harsh washing, retaining more non-specific bindings |
| Labeling of Neighboring Proteins | Labels non-interacting but neighboring proteins, providing insights into the proteomic landscape | Detects only direct interactions |
| Reducing False Positives/Negatives | Expresses fusion proteins in theoretically any cells, offering a more relevant context | Limited by specific conditions, with higher risks of false positives/negatives |
| Dynamic Process and Kinetic Data | Well-suited for studying spatially and temporally dynamic processes, providing kinetic data | Typically not designed for studying dynamic processes or generating kinetic data |
Significantly, it is worth noting that BioID biotinylation is a mark of potential proximity and not evidence for physical interactions. Thus, Creative Biolabs could also provide subsequent tests, like immunoprecipitation assays, to further validate proximity interactors identified by BioID and determine which reflect direct interactions in the physiological condition of cells.

Creative Biolabs provides state-of-the-art Proximity-dependent Biotin Identification (BioID) services, leveraging our extensive expertise in protein interaction mapping and cutting-edge biotinylation technology. With over 20 years of experience in molecular biology and protein analysis, our team tailors BioID strategies to meet specific research goals. By integrating proprietary tools and advanced biotinylation systems, we empower researchers in therapeutic target discovery, diagnostic biomarker development, and pathway elucidation.
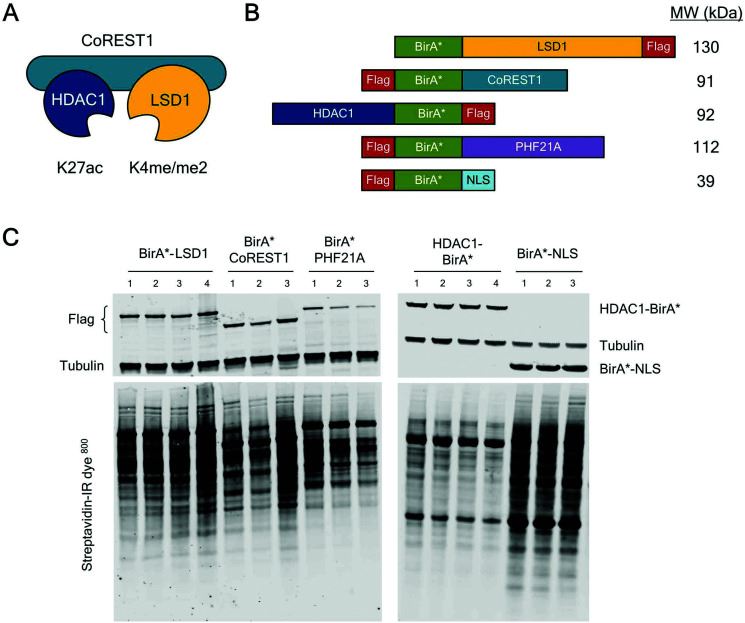 Fig. 3 CoREST complex members tagged with BirA* expressed in HEK293T cells biotinylated proximal proteins.3,6
Fig. 3 CoREST complex members tagged with BirA* expressed in HEK293T cells biotinylated proximal proteins.3,6
Lysine specific demethylase 1 (LSD1), as a part of the CoREST complex, regulates gene expression together with the REST co-repressor (CoREST) and histone deacetylase 1 (HDAC1). CoREST is recruited to specific genomic sites through a large number of transient interactions between core components, chromatin-related factors, and transcription factors. In this paper, using three different cell types, the researchers used proximity-dependent biotin identification (BioID) for four different members of the CoREST complex to recognize an integrated network of LSD1/CoREST-related proteins. In HEK293T cells, researchers identified 302 CoREST-related proteins. At the same time, they replaced endogenous LSD1 with BirA*-LSD1 in embryonic stem (ES) cells and performed BioID in pluripotent, early and late differentiation environments. This examines the dynamic properties of the CoREST interaction group in the primary cell type. Researchers also identified 156 LSD1-related proteins, 67 of which were constitutively associated at all three time points, meaning that most of the interacting proteins were dynamic and cell type dependent. Overall, researchers conducted 16 independent BioID assays on LSD1 using three different types of cells, producing a clear network of proteins associated with LSD1.
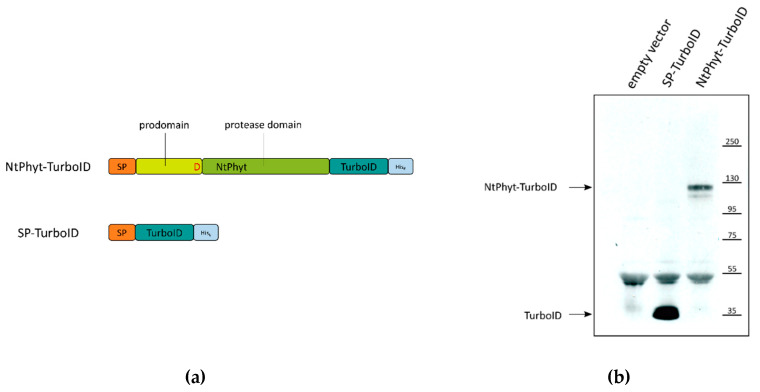 Fig. 4 Nicotiana tabacum phytaspase (NtPhyt)-TurboID and signal peptide (SP)-TurboID proteins utilized for the identification of phytaspase interactors.4,5
Fig. 4 Nicotiana tabacum phytaspase (NtPhyt)-TurboID and signal peptide (SP)-TurboID proteins utilized for the identification of phytaspase interactors.4,5
Because of their digestive activity and regulatory function, proteolytic enzymes play an important role in all aspects of plant development, including senescence. Here, researchers used a proximity-dependent biotin identification (BioID) assay to identify the protein chaperones of phytic acid proteases in Nicotiana benthamiana leaves producing phytaspase fused to a non-specific biotin ligase, TurboID. And several candidate protein interactions of phytic acid protease have been identified. These are mainly soluble residues of endoplasmic reticulum, namely endoplasmin, BiP, and calreticulin-3. For calreticulin-3, its gene is characterized by increased expression in senescent leaves, and its direct interaction with phytic acid protease was confirmed in the in vitro binding test using purified protein. In addition, a significant change in the post-translational modification of calreticulin-3 was observed in plant cells overproduced by phytic acid protease.
BioID is used in molecular biology to study protein interactions within living cells. It involves the use of a promiscuous biotin ligase that is fused to a protein of interest. This biotin ligase, when expressed in cells, catalyzes the biotinylation of nearby proteins, effectively labeling them. The biotinylated proteins can then be isolated and identified using streptavidin-based methods. This approach allows researchers to capture and identify proteins that are in close proximity to the protein of interest, providing insight into protein interaction networks and cellular organization.
Firstly, it does not require direct physical interactions between proteins, as it can identify proteins within a proximity of approximately 10 nanometers. This is particularly useful for detecting transient or weak interactions that are often missed by other methods. Additionally, BioID can be conducted under physiological conditions within living cells, providing a more accurate representation of protein interactions and dynamics in their native cellular context. Lastly, BioID is relatively straightforward and can be applied to a wide range of biological systems, making it a versatile tool for protein interaction studies.
BioID is particularly useful for exploring the molecular architecture of cells, understanding protein networks, and investigating cellular processes. It can help answer questions about the spatial organization of proteins within specific cellular compartments, identify novel components of protein complexes, or reveal proteins associated with particular cellular functions such as signaling pathways, organelle biogenesis, or disease mechanisms. By revealing proximity-based interactions, BioID can also assist in identifying potential targets for therapeutic intervention and provide insights into the molecular basis of diseases.
BioID has been adapted in various forms to enhance its versatility and specificity. For instance, a more rapid version called TurboID has been developed, which offers faster biotinylation speeds, allowing for shorter labeling times and reducing potential artifacts from overexpression and prolonged biotin ligase activity. Additionally, miniTurbo, a smaller version of TurboID, has been created to minimize the effects of the tag size on the function of the protein of interest. These adaptations make BioID suitable for dynamic studies where temporal resolution is crucial, such as monitoring changes in protein interactions during cell signaling events or other rapid cellular processes.
Originally developed for use in cell culture systems, BioID has been successfully adapted for use in tissue samples as well. This adaptation allows researchers to study protein interactions within more complex biological contexts, including multicellular environments and different organ systems. The application of BioID in tissues can provide insights into the spatial and temporal dynamics of protein networks in vivo, which is crucial for understanding physiological processes and disease states. However, applying BioID to tissues involves additional challenges, such as ensuring efficient delivery and expression of the biotin ligase fusion protein, and requires careful experimental design to obtain meaningful results.
BioID can significantly impact drug discovery by identifying novel targets and elucidating protein interaction networks that are critical for disease mechanisms. By mapping these interactions in disease states versus healthy states, researchers can identify key proteins that may serve as potential drug targets. Furthermore, BioID's ability to capture transient and weak interactions provides a more comprehensive understanding of the protein complexes involved in signaling pathways and disease progression, offering insights into the molecular underpinnings that can be leveraged for therapeutic intervention. This makes BioID an invaluable tool in the early stages of drug development, especially for diseases with complex pathologies such as cancer and neurodegenerative disorders.
BioID generally has a spatial resolution of about 10 nanometers, which is sufficient to identify proteins within the same protein complex or organelle. However, compared to other proximity labeling methods, BioID offers slightly better spatial precision. This finer resolution is advantageous for dissecting dense protein networks and can be critical in densely packed cellular environments, like synaptic junctions or the nuclear periphery.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
