Proximity-based labeling technique makes great contributions to protein-protein interaction detection and protein interactome network mapping. As an experienced services provider in pre-clinical research and drug discovery, Creative Biolabs has won a good reputation in completing the identification of protein-protein interactions. Based on the existing proximity-dependent biotin identification (BioID) service, our professional scientists here provide new TurboID services for protein-protein interaction exploration in vivo.
TurboID is an advanced enzyme-based proximity labeling (PL) technique developed to identify protein-protein interactions (PPIs) with improved efficiency and speed. It builds upon the BioID method, which uses a promiscuous mutant of the E. coli biotin ligase (BirA) to biotinylate nearby proteins. TurboID introduces engineered biotin ligases optimized through yeast display, significantly enhancing the labeling process.
 Fig. 1 Schematic representation of proximity-labeling systems.1,5
Fig. 1 Schematic representation of proximity-labeling systems.1,5
TurboID functions by tagging a protein of interest with a biotin ligase that catalyzes the attachment of biotin molecules to the lysine residues of neighboring proteins within approximately 10 nm. This biotinylation process creates a molecular "snapshot" of the protein's interaction environment. The biotinylated proteins are then purified using avidin-based affinity capture and identified through mass spectrometry (MS). Compared to traditional BioID, TurboID dramatically reduces the labeling time from 18-24 hours to as little as 10 minutes, making it more practical for dynamic or transient interaction studies.
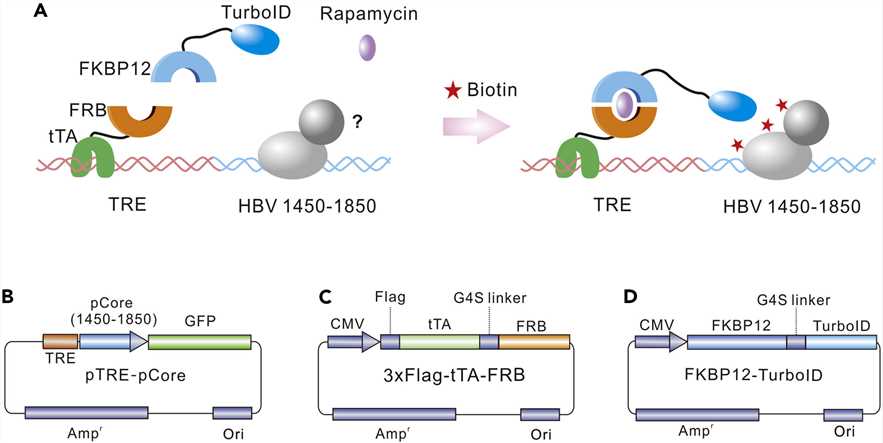 Fig.2 Establishment of a TurboID-based proximity labeling system.2,5
Fig.2 Establishment of a TurboID-based proximity labeling system.2,5
The combination of speed, sensitivity, specificity, and adaptability makes TurboID a powerful tool for studying protein-protein interactions and exploring complex biological systems.
| TurboID | Split-TurboID | BioID | BioID Variants (BioID2, BASU) | |
| Enzyme Type | Engineered biotin ligase | Split engineered biotin ligaseUse the resources in our library to help you understand your options and make critical decisions for your study. | Mutant E. coli biotin ligase (BirA R118G) |
BioID2: Aquifer aeolicus biotin ligase; BASU: Bacillus subtilis biotin ligase |
| Labeling Time | 10-30 minutes | ~1 hour (reconstitution-dependent) | 18-24 hours | BioID2: 3-6 hours; BASU: 1-2 hours |
| Catalytic Efficiency | High | Moderate | Low | Higher than BioID, but lower than TurboID |
| Proximity Radius | ~10 nm | ~10 nm | ~10 nm | ~10 nm |
| System Toxicity | Low | Low | Low | Low |
| Applications | General interactome studies, dynamic PPIs | Interaction studies of split complexes | Stable interactome studies | Similar to BioID with improved speed |
| Sensitivity | High | Moderate | Moderate | High |
| Specificity | High | High | Moderate | High |
| Key Advantages | Fast, efficient, non-toxic, highly versatile | Suitable for studying split proteins or weak interactions | Simple, robust for stable PPIs | Faster labeling compared to BioID |
| Limitations | Requires exogenous biotin in some systems | Requires split protein reconstitution | Long labeling time | Limited use in highly dynamic systems |
| Special Features | Designed for speed and efficiency | Studies spatially or temporally separated interactions | Cost-effective, easy setup | Compact size (BioID2), high activity (BASU) |
| Best Use Cases | Fast PPI studies, transient interaction mapping | Studies of protein-protein reconstitution | General PPI identification | Faster labeling in stable systems |
As a recognized biotech company in the field of drug discovery, Creative Biolabs has accumulated abundant expertise and experience in protein-protein interaction identifications. Based on the powerful technology platform, we are professional in providing one-stop and tailored solutions to advance PPI detection for our valued customers. In addition to our seasoned yeast two-hybrid (Y2H) and bacterial two-hybrid (B2H) services, here we successively launched BioID and TurboID to facilitate protein interaction research.

Creative Biolabs offers comprehensive TurboID-based Proximity Labeling Services, harnessing advanced biotinylation technology and extensive expertise in protein interaction mapping. TurboID's rapid and efficient labeling capabilities allow precise identification of protein-protein interactions, even in dynamic and transient systems. With cutting-edge tools and tailored strategies, we support research in therapeutic target discovery, biomarker identification, and pathway analysis, delivering high-quality results for complex interactome studies.
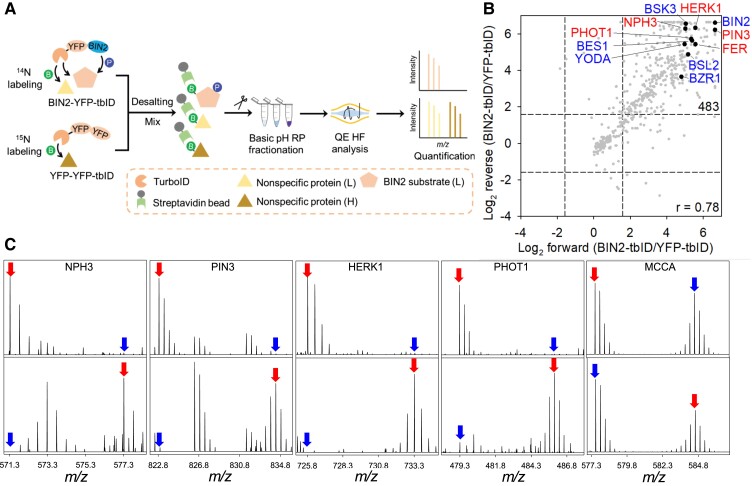 Fig. 3 TurboID-based identification of BIN2-proximal proteins in Arabidopsis.3,5
Fig. 3 TurboID-based identification of BIN2-proximal proteins in Arabidopsis.3,5
Since enzyme-substrate interactions are often short-lived, the enzyme-substrate relationship in post-translational modification (PTM) networks is often difficult to explain. Here, the researchers demonstrated that TurboID-based proximity labeling (TbPL) can effectively and specifically capture kinase and phosphatase substrates. More than 400 proximal proteins of the Arabidopsis thaliana BIN2 protein were identified by TbPL-mass spectrometry. Most of the proximal proteins of BIN2 show BIN2-dependent phosphorylation in vivo or in vitro. Protein-protein interaction network analysis showed that the proximal proteins of BIN2 included interactions with BIN2 substrates, which revealed a high level of interaction between BIN2 proximal proteins. The researchers established a BIN2 signaling network through proteomic analysis and revealed the role of BIN2 in regulating key cellular processes.
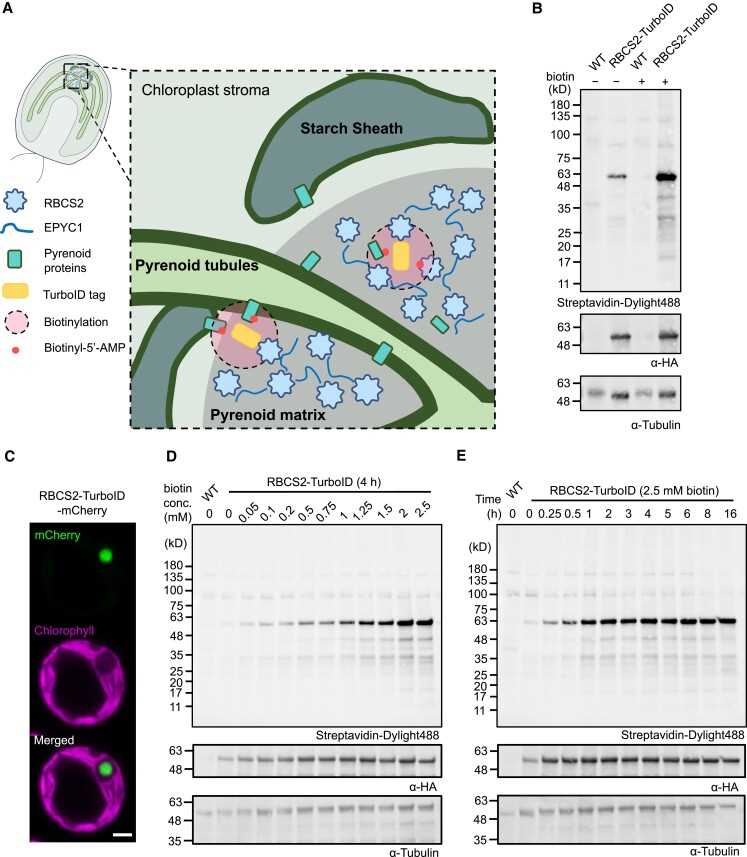 Fig. 4 Establishment and optimization of TurboID labeling in the Chlamydomonas chloroplast using RBCS2-TurboID lines.4,5
Fig. 4 Establishment and optimization of TurboID labeling in the Chlamydomonas chloroplast using RBCS2-TurboID lines.4,5
Phase separation is the basis of many biologically important cellular events, such as RNA metabolism, signal transduction and CO2 fixation. Traditional proteomics techniques limit the understanding and application of phase separation. In Chlamydomonas reinhardtii, Rubisco is condensed into a crucial phase-separated organelle called the pyrenoid that improves photosynthetic performance by supplying Rubisco with elevated concentrations of CO2. Here, the researchers developed a proximal labeling technique based on TurboID, in which the proximal proteins in Chlamydomonas chloroplasts can be labeled by biotin free radicals produced by TurboID labeling proteins. Then, by fusing the two core protein nuclear components with the TurboID tag, the scientists generated a nuclear proximal group containing most of the known nuclear proteins as well as new nuclear candidates. Fluorescent protein labeling of 7 previously uncharacterized TurboID recognition proteins showed that 6 of them were located in a series of subpyrenoid regions. The resulting proxiome also suggests that nuclear proteins have new secondary functions in RNA-related processes and redox-sensitive iron-sulfur cluster metabolism.
TurboID is an engineered biotin ligase that is used for proximity labeling in biological research. It is designed to be a faster and more efficient version of the protein labeling enzyme BioID. TurboID catalyzes the biotinylation of proteins that are near it in living cells and tissues. This process involves the enzyme facilitating the addition of a biotin tag to lysine residues on proteins that are within approximately 10 nanometers of TurboID. This proximity labeling is particularly useful for studying protein interactions and the composition of protein complexes in their native cellular environment. The enhanced speed and efficiency of TurboID allow for shorter labeling times, ranging from minutes to a few hours, which is significantly faster compared to the original BioID that may require up to 24 hours.
TurboID finds extensive applications in molecular biology and biochemistry, particularly in the study of protein-protein interactions and the dynamics of protein networks within cells. It is especially valuable in identifying transient or weak protein interactions that are difficult to detect with traditional methods. Researchers use TurboID for various applications including mapping the components of specific cellular organelles, understanding signal transduction pathways, and exploring the changes in protein interactions under different physiological conditions or in disease states. Additionally, due to its effectiveness in living cells and organisms, TurboID is also applied in in vivo studies, providing insights into the biological processes at the organismal level.
TurboID offers several advantages over traditional proximity labeling methods such as BioID or APEX. First, it requires much shorter incubation times to achieve effective labeling, ranging from 10 minutes to an hour, compared to the 24-hour incubation period often necessary for BioID. This rapid action reduces potential perturbations to cellular processes and minimizes the effects of overexpression toxicity. Secondly, TurboID demonstrates enhanced biotinylation efficiency, which improves the detection of low-abundance or transient protein interactions. Moreover, it functions efficiently at lower temperatures and in a broader range of biological systems, including less permeable cells and tissues, making it versatile for various experimental setups.
One primary challenge is the potential for background labeling, where non-specific biotinylation can occur due to the high activity of the enzyme. This can lead to false positives in protein interaction studies. Additionally, the high efficiency of TurboID might result in over-biotinylation, which could mask or alter the function of target proteins. Researchers must optimize experimental conditions carefully to balance labeling efficiency with specificity. Furthermore, like all biotin-based methods, TurboID requires the availability of free biotin in the cell, and its efficiency can be affected by biotin competition or depletion in the cellular environment.
Yes, TurboID can be effectively used in live animal models, expanding its utility beyond cell culture systems. This capability is particularly beneficial for in vivo studies where understanding protein interactions and dynamics within the actual physiological context is crucial. For example, researchers can express TurboID in specific tissues or cells of model organisms like mice through genetic engineering techniques. By administering biotin systemically or locally, proteins proximal to TurboID in these living organisms can be labeled and later analyzed to gain insights into tissue-specific or developmental processes.
First, the concentration of biotin available in the cell or tissue environment needs to be carefully controlled, as insufficient biotin can lead to reduced labeling efficiency while excessive biotin might increase background noise. The expression level of TurboID also needs to be optimized to avoid toxicity and ensure it does not interfere with normal cellular processes. Additionally, the choice of the fusion strategy (whether TurboID is fused to a specific protein of interest or expressed alone) can affect the proximity labeling results. Proper controls should be established to differentiate between specific interactions and background labeling, such as using cells expressing TurboID without biotin or with a catalytically inactive version of the enzyme. Lastly, due to the rapid action of TurboID, timing the biotin administration correctly is crucial to capturing the dynamic changes in protein interactions, especially in studies involving transient cellular events or stimuli.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
