Yeast hybrid systems, especially yeast two-hybrid (Y2H), have been recognized as one of the most standardized in vivo approaches for protein-protein and protein-nucleic acid interaction screening and detection. Experienced in providing protein-protein interaction assay services, Creative Biolabs has developed and launched drug-target interaction services based on our perfect Y2H technology platform to boost novel drug discovery.
Almost all drugs, no matter small molecule compounds or biological macromolecules, achieve their therapeutic effects through binding to the specific target proteins and thus affecting the biological activities or functions of the targets. The identification of drug-target interactions is a crucial factor in the drug discovery process, which not only contributes to the explication of the pathomechanism of diseases and mechanisms of drugs but also aids to maximize the therapeutic effect while reducing adverse side effects.
Drug-target interaction screening is a time-consuming, laborious, and complex process, which is roughly divided into computational prediction and experimental validation. The computational/in silico technique is an important, effective, and time & cost-saving approach to predict drug-target interactions on a large scale. Drug-target interaction prediction by in silico technique then should be performed and validated by biological assays and animal experiments. Currently, a variety of in vivo and in vitro experiments have been developed to screen and identify drug-target interactions, such as protein microarray, pull-down assay, affinity chromatography, yeast hybrid assays, and so forth.
In 1989, yeast two-hybrid (Y2H) system has been introduced to screen and detect protein-protein and protein-nucleic acid interactions in the yeast Saccharomyces cerevisiae. It is based on the principle that the Gal4 transcription factor in the yeast Saccharomyces cerevisiae is composed of two functional domains, one is the DNA-binding domain mediating DNA binding and the other is the activation domain responsible for transcription activation. The two functional domains of the Gal4 transcriptional activator can not activate the transcription when they are separated. Only when the two separated passages are relatively close in space, the complete GAL4 transcription factor activity can be represented, and the downstream promoter of the upstream activation sequence can be activated, so that the gene downstream of the promoter can be transcribed. Over these decades, Y2H has been extensively applied for protein interaction screening and identification all through the world, and different Y2H variant methods also have been developed.
 Fig.1 Y2H methods and their applications to detect protein-protein or protein-small-molecule interactions. (Hamdi, 2012)
Fig.1 Y2H methods and their applications to detect protein-protein or protein-small-molecule interactions. (Hamdi, 2012)
In order to the excellent performance in the detection of protein-protein and protein-nucleic acid interactions, Y2H also is a valuable tool for chemical drug-target interaction identification. In Y2H-based drug-target interaction screening or identification, a tool protein is introduced to be fused with the DNA-binding domain and tagged chemical compounds or drugs of interest then bind to the tool protein. The whole complex is used as a bait to screen the RNA, cells, or tissues prepared library on the activation domain. Also, this system can be used for the validation of the interaction of the drug with the specified protein/target one by one.
 Fig.2 Drug-target interaction identification or screening based on the Y2H system. (Creative Biolabs)
Fig.2 Drug-target interaction identification or screening based on the Y2H system. (Creative Biolabs)
Creative Biolabs has completed plenty of protein-protein interaction screening or validation based on yeast hybrid systems, accumulating abundant expertise and experience in protein-protein interaction identifications. Added by rich experience and the perfect technology platform, our seasoned scientists offer one-stop or tailored Y2H-based drug-target interaction identification services to global clients.
Just feel free to contact us and communicate with us about your questions, our experienced technicians will give you the most tailored solutions to you.
Other optional protein-protein interaction (PPI) assay services:
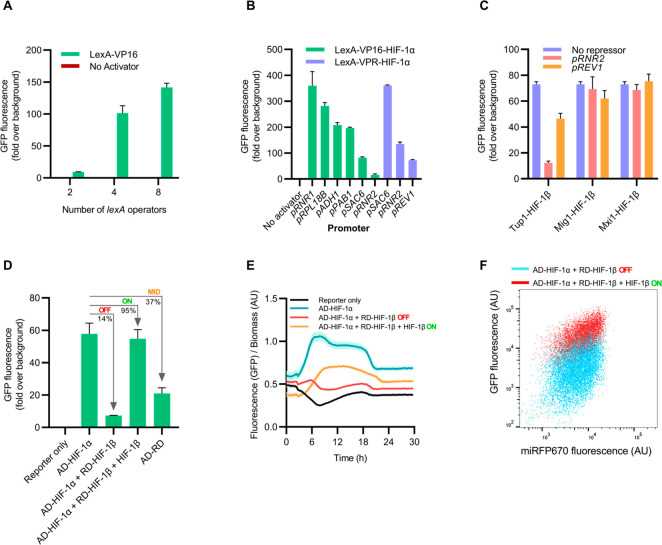 Fig. 3 Design and construction of a protein–protein interaction biosensor with fluorescence output. (Louis H. Scott, 2022)
Fig. 3 Design and construction of a protein–protein interaction biosensor with fluorescence output. (Louis H. Scott, 2022)
The heterodimeric transcription factor, hypoxia inducible factor-1 (HIF-1), is an important anticancer target. Here, the researchers optimized the yeast two-hybrid system for suppressing transcriptional activators and verified it with HIF-1. Under the selection conditions based on fluorescence and nutritional defects, the yeast two-hybrid system can detect the inhibition of HIF-1α/HIF-1β dimerization. In addition, they designed mechanisms to control cell activity and the effects of off-target drugs. Through the above systems and mechanisms, scientists have characterized all parts of the biosensor system and believe that the tool will be generally applicable to a wide range of protein-protein interaction targets. The researchers expect the biosensor to play a role in screening new models of drugs encoded by DNA.
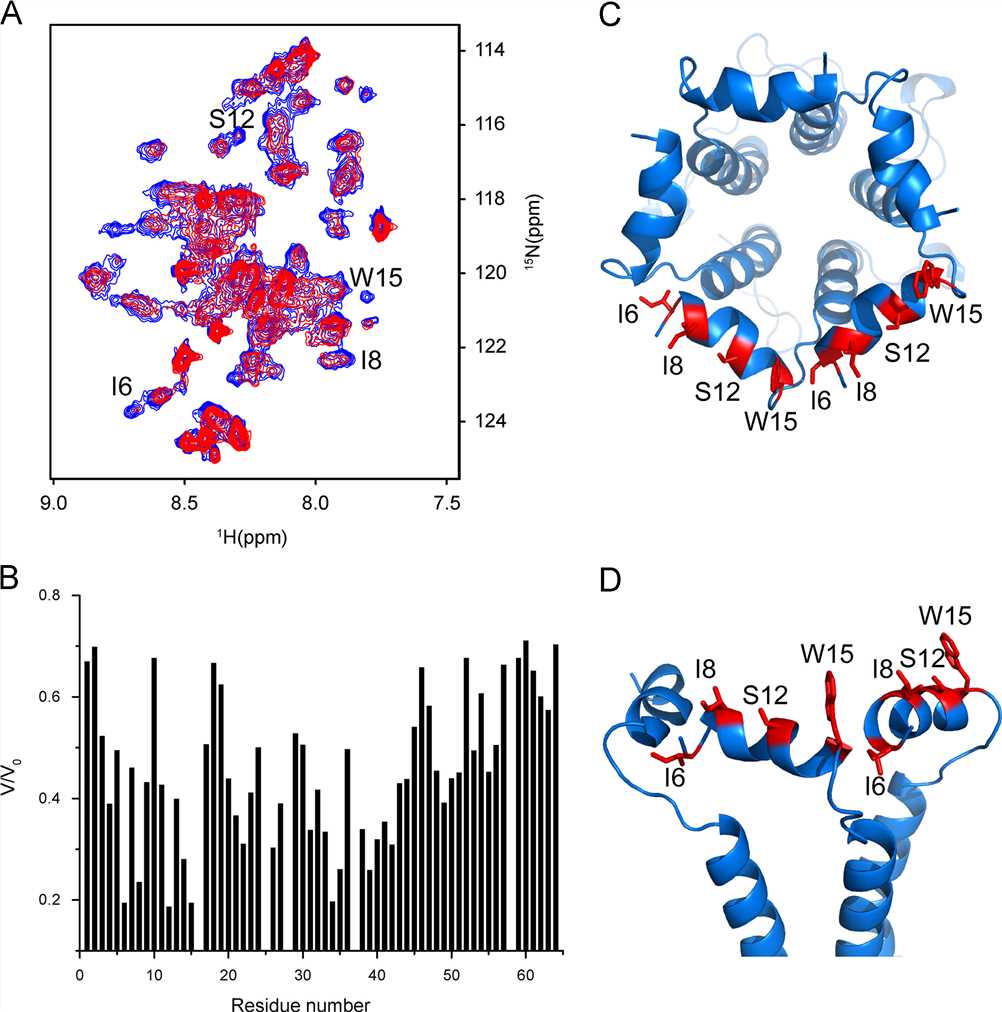 Fig. 4 Interaction between SH protein and BAP31. (Yan Li, 2015)
Fig. 4 Interaction between SH protein and BAP31. (Yan Li, 2015)
Small hydrophobic (SH) protein is a short channel-forming peptide encoded by the human respiratory syncytial virus (hRSV). The absence of SH protein can weaken the virus in mice and primates and delay the apoptosis of infected cells. The researchers used a membrane-based yeast two-hybrid system (MbY2H) and a library from human lung cDNA to detect proteins that bind to SH proteins. Through the above experiments, the researchers identified a membrane protein, B cell associated protein 31 (BAP31). The transfected SH protein and the transfected BAP31 co-located in the cells, and the endogenous BAP31 was pulled down. The C-terminal inner domain of purified SH was titrated with isotope-labeled BAP31 proteins in detergent micelles. The results showed that there was a direct interaction between the two proteins. Given that BAP31 plays a key role in protein transport and pro- and anti-apoptotic pathways, this new interaction may constitute a potential drug target.
Yeast two-hybrid (Y2H) based drug-target interaction identification is a technique used in molecular biology and pharmacology to discover and confirm the interactions between a drug and its potential target proteins. This method utilizes a genetic system in yeast cells that can report the interaction between two proteins by activating the transcription of a reporter gene, which is only possible when the two proteins bind to each other. In the context of drug discovery, one of the proteins is typically a drug or a small molecule linked to a binding domain, and the other is a potential target protein. When the drug binds to the target protein, the reporter gene is activated, indicating a successful interaction. This method is invaluable for validating the mechanisms of new drugs and understanding their effects at the molecular level.
Y2H-based drug-target interaction identification can efficiently and accurately identify the molecular targets of new drugs. This information is essential for understanding the therapeutic potential of a drug, predicting its side effects, and optimizing its efficacy. By confirming how and with which proteins a drug interacts within the cell, scientists can better design drugs that are more specific to their target, potentially reducing unintended interactions that could lead to adverse effects. Furthermore, this technique can be used to identify off-target effects, helping in the refinement of drug candidates to ensure that they are safe and effective before proceeding to clinical trials.
The Y2H-based drug-target interaction identification process involves several key steps. Initially, the drug or small molecule of interest is chemically linked to a DNA-binding domain, which is part of a transcription factor needed to activate a reporter gene. This hybrid is then introduced into yeast cells along with a library of target proteins, each fused to an activation domain of the transcription factor. If the drug-bound DNA-binding domain interacts with a target protein fused to the activation domain, the two halves of the transcription factor come together to form a functional unit that can initiate transcription of the reporter gene. The activation of this reporter gene indicates a positive interaction between the drug and the target protein, which can then be further validated through additional experiments.
Y2H-based drug-target interaction identification is versatile, but it has some limitations in terms of the types of drugs that can be effectively studied using this method. Primarily, the technique is suitable for small molecules and peptides that can penetrate the yeast cell and reach the nucleus where the interaction detection system is active. However, larger molecules such as biologics, including large peptides, proteins, and antibodies, may not be suitable for Y2H screens because they cannot easily enter yeast cells or because their size and complexity might prevent proper interaction within the confines of the nuclear environment. Additionally, drugs that require specific cellular contexts or post-translational modifications for activity may not be accurately assessed using Y2H.
To address the limitations of traditional Y2H systems, several enhancements and alternative strategies have been developed. One such improvement is the incorporation of membrane yeast two-hybrid (MYTH) technology, which adapts the Y2H system to detect interactions at the membrane level, allowing the identification of drug interactions with membrane proteins. Additionally, modifications to the yeast expression system, such as the use of humanized yeast strains that can perform post-translational modifications similar to human cells, have expanded the range of potential drug-target interactions that can be studied. Advanced cloning techniques and high-throughput screening methods have also improved the efficiency and accuracy of Y2H screens. Furthermore, integrating Y2H data with other biochemical and biophysical methods in a multi-tiered screening approach helps to validate and refine potential drug-target interactions, enhancing the overall robustness of drug discovery programs.
One of the primary benefits is its high-throughput capability, allowing researchers to screen thousands of potential interactions quickly and cost-effectively. This method is also highly versatile, capable of identifying interactions between a wide variety of protein classes and small molecules, provided they can be engineered to enter the yeast nucleus. Additionally, Y2H is relatively simple to set up and does not require complex instrumentation, making it accessible to many laboratories. The genetic nature of the readout, which relies on transcriptional activation, allows for easy detection and quantification of interactions. This method is particularly useful for identifying novel targets that may not have been predicted by other methods, thereby expanding the potential therapeutic applications of new drugs.
Typically, interactions identified by Y2H are first retested within the system to rule out false positives due to experimental anomalies. Subsequent validations involve a variety of biochemical and cellular techniques. For instance, co-immunoprecipitation (co-IP) and pull-down assays are used to confirm the physical interactions between the drug and the target protein in a more controlled environment. Additionally, functional assays, such as reporter assays in mammalian cells, can be employed to assess whether the interaction affects the biological activity of the target protein. Further, techniques like surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC) provide quantitative measures of the binding affinity and kinetics of the interaction. Ultimately, in vivo studies in model organisms or cell lines relevant to the disease being targeted are conducted to establish the biological significance of these interactions in a physiological context.
Y2H-based drug-target interaction identification can reveal direct interactions with specific proteins within a cellular environment. Understanding which proteins a drug interacts with can help to infer its biological pathways and potential effects on cellular processes. For example, if a drug is found to interact with a protein that regulates cell cycle progression, it might suggest that the drug has potential applications in treating diseases characterized by uncontrolled cell division, such as cancer. Additionally, identifying unintended interactions can provide insights into possible side effects or off-target effects, which are crucial for drug safety assessments. This mechanistic understanding is essential for refining drug design, optimizing dosing strategies, and predicting potential drug-drug interactions in multi-drug therapies.
One area of ongoing development is the integration of Y2H systems with next-generation sequencing technologies, which would allow for even more high-throughput and comprehensive screening capabilities. This integration can facilitate rapid sequencing of large numbers of clones to identify interactions without the need for individual colony picking, dramatically speeding up the process. Additionally, efforts are being made to improve the sensitivity and specificity of Y2H systems by incorporating more sophisticated reporter genes and developing better vectors that minimize background activation and false positives. Another exciting prospect is the adaptation of Y2H systems to more closely mimic human cell conditions, potentially through the use of humanized yeast or by incorporating human cell extracts, enhancing the clinical relevance of the interactions discovered. These advancements will further solidify the role of Y2H in drug discovery and development processes.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
