Creative Biolabs supplies highly sensitive Protein-fragment Complementation Assay (PCA) service to detect dynamic protein-protein interactions (PPIs), conformation changes, and protein complex dimensions in virtually any cell type or organism.
In PCA, protein A and B, two proteins of interest, is fused to one complementary fragment of a rationally dissected reporter protein, respectively. If the protein A and B could interact, the reporter fragments are taken together, fold and reconstitute its native structural activity. The reporter proteins have been picked as those generating a detectable signal, including fluorescent, luminescent, colorimetric signal, and survival selection assay. Owing to its high sensitivity, high signal-to-background ratio, as well as full reversibility, PCA is an extremely powerful and widely used approach to monitor direct and proximal associations between proteins expressed at endogenous level in any intact living cell, in subcellular compartments, in vivo or in vitro. The reversibility of PCA assures accurate measurements of the kinetics and equilibrium aspects of PPI assembly or disassembly.
In addition to the above mentioned, Creative Biolabs also displays the following capabilities of PCA: the detection of induced versus constitutive PPIs under any conditions (including occurrence in hormone, nutritional, developmental or environmental-induced signals) and screening of cDNA libraries for PPIs in any cell type.
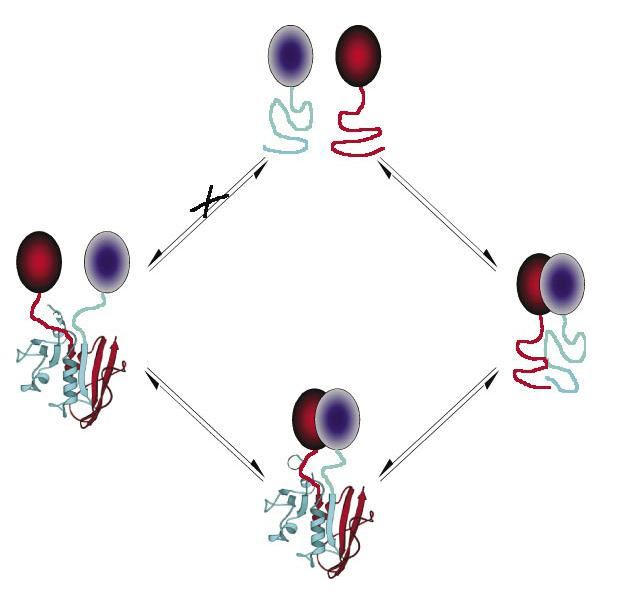 Fig.1 The protein-fragment complementation assay (PCA) strategy.1
Fig.1 The protein-fragment complementation assay (PCA) strategy.1
PCA acts as a general experimental method that enables the quantitative detection of dynamic PPIs and provides structural and topological details of how a PPI is formed or if complexes go through conformation changes. From above chart, a crucial point of this approach is that the fragments are designed so that they can’t fold spontaneously; otherwise a situation would occur that results in a false-positive signal.
In the process of PCA, PPIs are measured depending on fusing each of the proteins of interest to complementary N- or C-terminal peptides of a reporter protein that has been rationally dissected. The split reporter proteins are brought into proximity after the interaction of two interest proteins, allowing them to fold into the three-dimensional structure of the reporter protein, thus reconstituting the activity of the reporter signal.
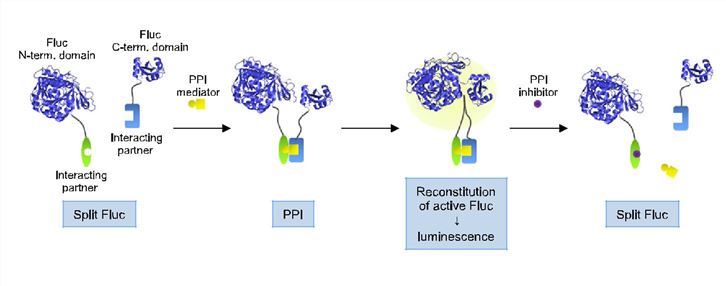 Fig.2 Principle of Fluc PCA.2
Fig.2 Principle of Fluc PCA.2
This general principle of PCA has been applied to generate assays using reporter proteins, whose activities are measured in a number of different ways, depending on the desired applications.
Typical PCA has been using dihydrofolate reductase (DHFR), β-lactamase, GFP or luciferase producing a detectable signal in split protein PCAs. When fluorescent proteins are reconstituted the PCA is called bimolecular fluorescence complementation assay (BiFC).
The DHFR-PCA can be used in numerous applications to display both simple survival-selection as readout and a fluorescent assay simultaneously allowing quantitative detection and cellular localization of protein interactions to be performed. Unlike DHFR, the β-lactamase-based PCA assay is worked as a much sensitive in vivo or in vitro quantitative detector of PPIs as one way measures the continuous conversion of substrate to fluorescent or colored product.
Creative Biolabs has completed a growing number of study services using PCA, which demonstrates the broad potential of this approach. The PCA technique not only can analyze the localization of protein molecular interaction dynamically, map intracellular signal transduction pathway and protein biochemical network but also is applied to protein library, target discovery and high throughput drug screening.
Other optional Protein-Protein Interaction (PPI) Assay services:
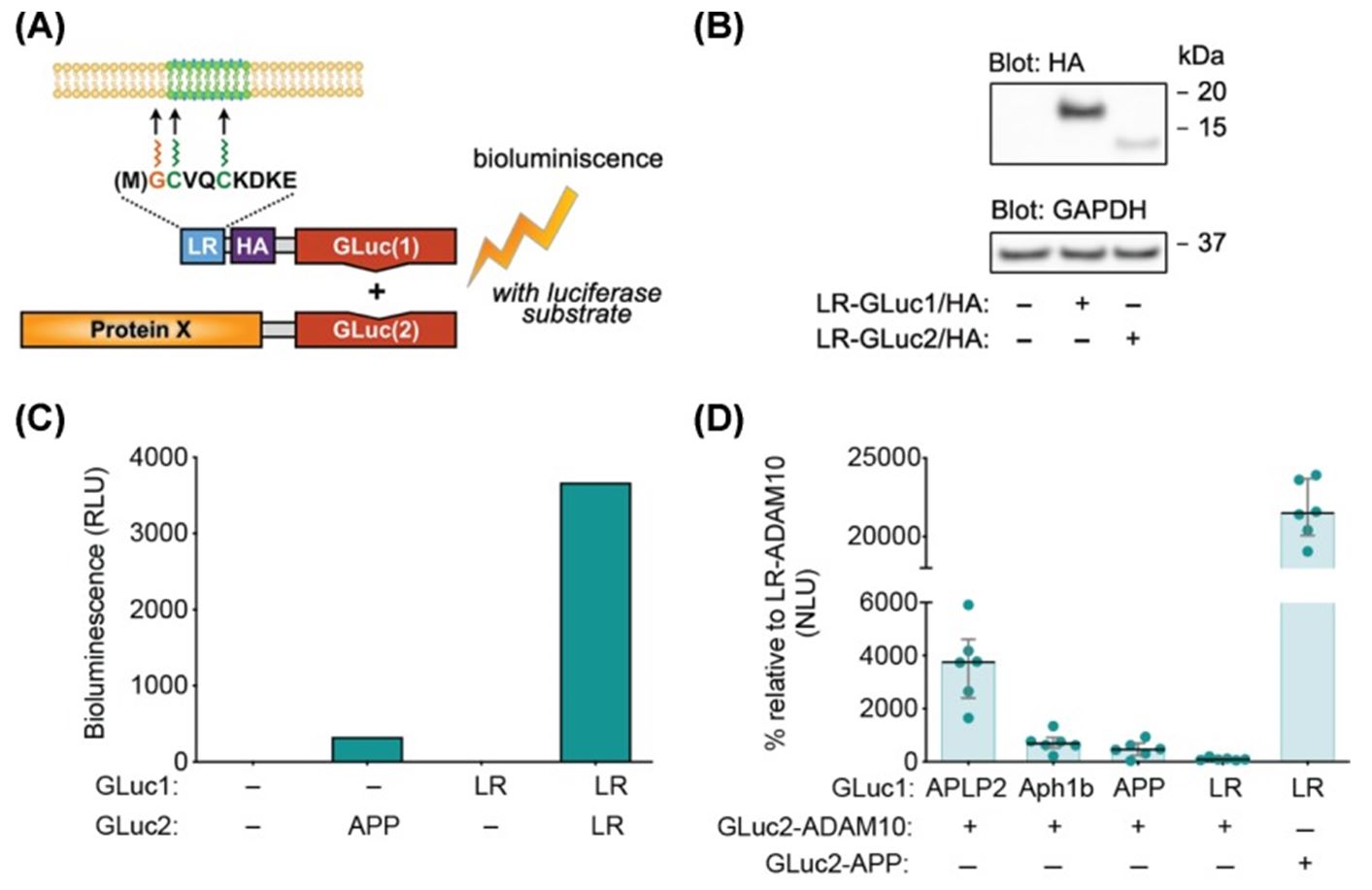 Fig.3 Graphical presentation of live-cell membrane raft-localization reporter assay (LR-PCA).3
Fig.3 Graphical presentation of live-cell membrane raft-localization reporter assay (LR-PCA).3
The plasma membrane consists of a variety of discrete domains, whose composition and characteristics are different from those of the surrounding membrane. The selective allocation of proteins to these discrete domains is essential for the function and integrity of the membrane. In this paper, the researchers developed a new method based on the Gaussia princeps luciferase protein fragment complementarity assay to quantitatively study the distribution of proteins to lipid rafts in intact living cells. In the assay, the reporter construct, carrying one half of the luciferase protein, is targeted to lipid microdomains through the fused acetylation motif from the Src-family kinase Fyn. The target protein carries the other half of the luciferase protein. Together, the two can be used as reversible real-time sensors for protein raft recruitment. This assay has been proven to be able to detect the dynamic changes in the raft localization of two key proteins, Akt and APP. At the same time, this method can be used for high-throughput screening and the large-scale study of other living cells.
The protein-fragment complementation assay (PCA) is a biochemical technique used to study protein-protein interactions. It works by splitting a reporter protein into two separate fragments, each of which is fused to one of the proteins of interest. These protein fragments are non-functional when apart but can reconstitute the active reporter protein when the fused proteins of interest interact. This reconstitution brings the fragments close enough to restore the structural integrity of the reporter protein, allowing it to function normally. The restoration of function, which could be enzymatic activity or fluorescence, provides a measurable indication of the interaction between the target proteins.
PCA is widely used in molecular biology and biochemistry for investigating interactions between proteins within living cells. This method is particularly useful for confirming interactions identified through other methods like yeast two-hybrid assays. It is also employed in drug discovery and development processes to identify and validate potential drug targets. One of the primary advantages of PCA is its ability to detect and measure interactions in their native cellular environment, which can provide more relevant biological insights. Additionally, PCA can be adapted for use in various types of cells and organisms, enhancing its versatility as a research tool.
Luciferase: Used for its bioluminescent properties, allowing real-time monitoring of protein interactions.
Green fluorescent protein (GFP) and its variants: These fluorescent proteins enable the visualization of protein interactions using fluorescence microscopy.
Β-galactosidase: Often used in bacterial systems, this enzyme allows for colorimetric read-outs based on its activity, which is easy to measure and quantify.
Dihydrofolate reductase (DHFR): This enzyme is used for its ability to confer cell survival in media lacking certain nutrients, making it useful for selection assays.
PCA has been adapted for high-throughput screening (HTS) applications, which allows researchers to systematically study large sets of protein interactions or screen for modulators of these interactions in a scalable manner. For high-throughput setups, PCA is often integrated with automated liquid handling systems and readout devices that can measure luminescence or fluorescence. The assays are typically conducted in multi-well plates, such as 96-well or 384-well formats, enabling the simultaneous testing of thousands of conditions or samples. This adaptation is particularly useful in drug discovery, where PCA can be employed to identify small molecule inhibitors or enhancers of protein-protein interactions.
Yes, PCA can be used in vivo to study protein interactions within the context of a living organism. This application is crucial for understanding the physiological roles of protein interactions. To conduct PCA in vivo, the genes encoding the fusion proteins are introduced into the organism using vectors suitable for transgenic expression, such as viral vectors or plasmids. Commonly used organisms for in vivo PCA include mice, zebrafish, and drosophila. The readouts in such experiments may involve whole-body imaging of luminescent or fluorescent signals, providing insights into where and when specific protein interactions occur within the organism.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
