With state-of-the-art facilities and enriched experience in antibody engineering, Creative Biolabs has developed excellent CreMap™ epitope mapping & discovery platform to provide customers professional technical services in a time- and cost-efficient manner.
Epitope, or antigenic determinant, is typically a segment on an antigen that can be recognized by antibodies or T cells for triggering the specific immune responses. Epitope mapping is the process of identifying the binding sites of antibodies (or TCRs) on target antigens, which helps discover the new therapeutics, vaccines, and diagnostics. Identifying epitopes also gives a clue about the mechanism of binding to antibodies or T cell receptors. In most cases, epitope mapping is performed with monoclonal antibodies (mAbs).
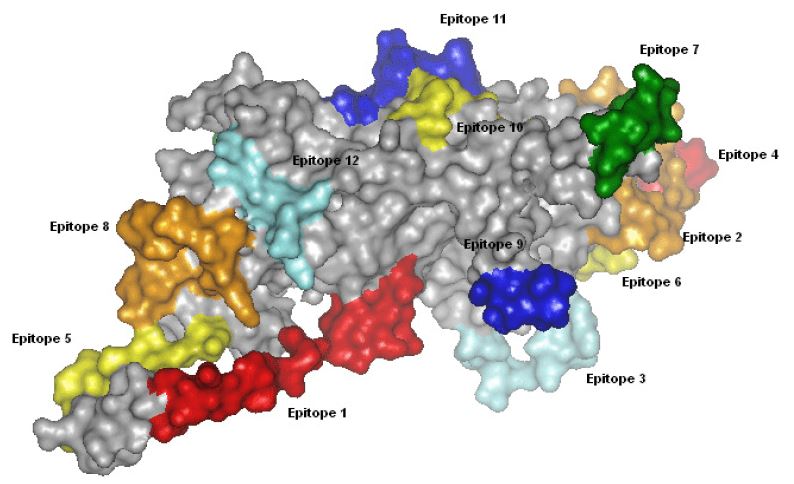 Fig.1 Predicted B cell epitopes of Human papillomavirus type 16 major late capsid protein L1.
Fig.1 Predicted B cell epitopes of Human papillomavirus type 16 major late capsid protein L1.
The techniques determining the potent epitopes (both T cell epitopes and B cell epitopes) have been greatly improved by the application and development in immunology for the past years. And these robust techniques could help you to:
Creative Biolabs' outstanding research group has developed the CreMap™ B Cell Epitope Mapping Platform and CreMap™ T cell Epitope Mapping Platform based on the professional knowledge and enriched experiences. We provide excellent epitope mapping services for our clients based on our advanced platforms.
The B cell epitopes are the binding sites of an antigen recognized by the corresponding antibodies. Based on the different binding characteristics, B cell epitopes contain two categories: linear epitopes and conformation epitopes. The linear epitopes are continuous sequences of amino acid on peptide chains, whereas the conformation epitopes act as sequence discontinuous amino acid chains but spatially adjacent.
Our expert group in Creative Biolabs has developed an advanced B cell epitope mapping platform, which contains several advanced technologies for mapping both conformational and linear epitopes in a high-throughput and efficient way. Read more…
In the CreMap™ T cell epitope discovery platform, Creative Biolabs has developed an exceptional MHC-peptide binding assay for determining the potent binding sites of the T cell with effective approaches. Compared with other ineffective and time-consume strategies, this platform provides a cost-effective solution for your epitope detection when you are attempting to design a peptide vaccine or develop a protein drug to certain diseases. Read more…
Over the past few years, Creative Biolabs has been providing best-in-class epitope mapping service by our excellent CreMap™ platform, aiming to guide your epitope mapping and discovery research. If you are interested in our service, please contact us for more detail information.
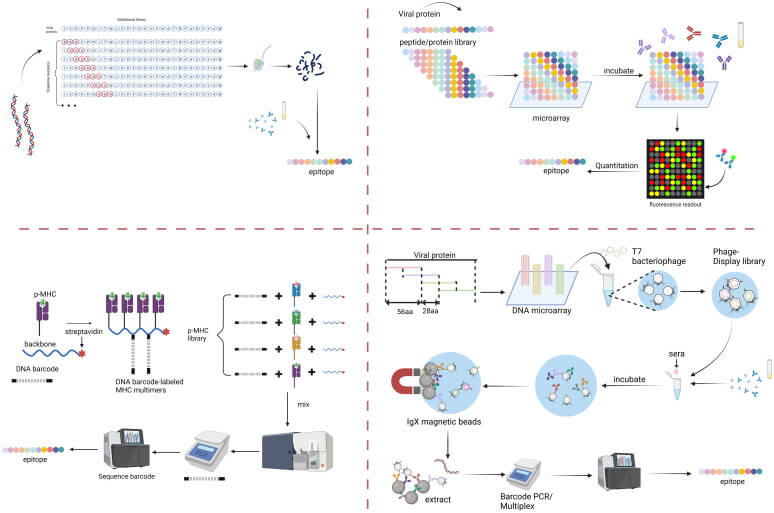 Fig. 2 Multiplexed serology for viral epitope mapping. (Diya Hu, 2023)
Fig. 2 Multiplexed serology for viral epitope mapping. (Diya Hu, 2023)
The article explores the research significance of massively multiplexed epitope mapping techniques for viral antigen discovery, emphasizing their application in immunotherapy and vaccine development. Epitope mapping's main goal is to identify the minimal regions on antigens that immune cells recognize, which is crucial for developing effective vaccines and therapies against viruses. The study demonstrates the use of high-throughput technologies, such as bacteriophage-display libraries and deep mutational scanning, to identify and characterize viral epitopes comprehensively. These methods allow for the rapid screening of a vast number of epitopes, aiding in the quick response to viral mutations and the design of targeted immunotherapies.
Epitope mapping is the process of identifying the specific parts, or epitopes, of antigens that are recognized by immune cells, such as antibodies or T-cell receptors. This technique is crucial for understanding how pathogens interact with the immune system, designing vaccines, and developing diagnostic tests.
Several techniques are used for epitope mapping, including X-ray crystallography, nuclear magnetic resonance (NMR), mass spectrometry, phage display, yeast display, and computational prediction models. These methods help reveal the structural and functional characteristics of epitopes.
Phage display involves expressing peptide libraries on the surface of bacteriophages, which are then screened against antibodies or T-cell receptors to identify binding interactions. This technique is highly versatile and allows for the screening of millions of peptides, making it a powerful tool for epitope discovery.
Epitope mapping can be used to enhance the effectiveness of existing vaccines by identifying new, more potent or broadly reactive epitopes. This information can be used to update vaccine formulations to cover emerging pathogen variants or to improve immune responses in different populations.
Computational biology plays a crucial role in epitope mapping by using algorithms and models to predict epitope structures and their interactions with antibodies or T-cell receptors. These tools can significantly speed up the discovery process and reduce the need for extensive laboratory testing.
B-cell epitopes are often mapped using methods that identify continuous or conformational epitopes on the surface of proteins, such as phage display or mass spectrometry. T-cell epitopes, which must be presented by MHC molecules, are often mapped using peptide-MHC multimer technologies or cellular assays that measure T-cell responses.
Epitope mapping faces several challenges, including the complexity of protein structures, the vast diversity of epitopes, and the difficulty of predicting which epitopes will elicit a protective immune response. Additionally, technical limitations in accurately mimicking the natural processing and presentation of epitopes can affect the accuracy of mapping.
In personalized medicine, epitope mapping can be used to tailor immunotherapies to individual patients, especially in cancer treatment. By identifying tumor-specific epitopes, therapies can be designed to target only cancer cells, minimizing damage to normal tissues and improving treatment outcomes.
Future advancements in epitope mapping are likely to come from improvements in high-throughput screening technologies, more accurate computational models, and better understanding of the immune system's interaction with different epitopes. These developments will enhance the speed and precision of vaccine design and immunotherapy development.
Use the resources in our library to help you understand your options and make critical decisions for your study.
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.
| USA:
Europe: Germany: |
|
|
Call us at: USA: UK: Germany: |
|
|
Fax:
|
|
| Email: info@creative-biolabs.com |
